rmultinom (10, 1, c(.4,.1,.2,.3))
## [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10]
## [1,] 0 0 1 0 1 0 1 0 1 0
## [2,] 0 0 0 0 0 1 0 1 0 0
## [3,] 0 0 0 1 0 0 0 0 0 1
## [4,] 1 1 0 0 0 0 0 0 0 012 Multinomial and Ordinal regression
In Chapter 10 we introduced models that can predict dichotomous (2-category) categorical variables using logistic regression. Here, we extend the concepts introduced in that chapter to the modeling of dependent variables with any number of categories. First, we discuss multinomial regression models that can be used to predict categorical variables without an inherent ordering. For example, we could use a multinomial model to predict the perception of vowel sounds from speech acoustics, to predict the lexical category of a word (verb, noun, adjective, …), or to predict a speaker’s native language. After this, we introduce ordinal models appropriate for the prediction of categorical variables with some inherent ordering (e.g. first, second, and third).
12.1 Chapter pre-cap
In this chapter, we introduce models for the prediction of categorical dependent variables with >2 categories. First, we discuss multinomial regression and its relationship to logistic regression. A multinomial logistic regression model with several predictors and random effects is fit and interpreted. We then explain the use of multinomial models to classify data, model participant responses, and make territorial maps. After that, we introduce ordinal logistic regression models, models to predict ordered categorical data. We present ordinal regression as a latent variable model, and discuss the use of cumulative distributions and thresholds by ordinal models. An ordinal logistic regression model with several predictors and random effects is fit, interpreted, and discussed. Finally, we present an example of an ordinal regression model with participant-specific latent variable distributions.
12.2 Multinomial logistic regression
Multinomial logistic regression allows you to model data generated by a multinomial distribution with unknown parameters. Multinomial data arises when you have a variable that can take on any number of discrete values (often, but not always, a small number). These values do not need to have a necessary order (but they can), and the ‘differences’ between them don’t mean anything. For example, in our experiment listeners were asked to identify speakers as either a boy, a girl, a man, or a woman. There was no choice but to respond one of these values, there is nothing ‘between’ them, and it doesn’t make sense to talk about, e.g. boy plus girl or woman minus man. Further, although these categories could potentially be ordered in several ways, no ordering is inherent to the categories.
The multinomial distribution is a generalization of the binomial distribution (see Section 10.2) to any number of discrete categories. This distribution will allow us to model the categorization of speakers into boys, girls, men, and women simultaneously rather than modeling this as two binary categorizations (male vs. female and adult vs. child). The multinomial distribution has three parameters:
- \(n\): The number of trials, a positive integer.
- \(J\): The number of possible outcome categories, a positive integer.
- \(p_1, \dots, p_J\): A vector of probabilities of observing each of the \(J\) outcomes where \(\sum p_j = 1\).
The multinomial distribution (equation Equation 12.1) takes the above parameters and generates a vector of counts (\(y_1, \dots, y_J\)) representing the number of observations for each category. For every observation of a multinomial variable, the sum of the outcome probabilities (\(p_1, \dots, p_J\)) must equal 1 and the sum of the outcomes (\(y_1, \dots, y_J\)) must equal n, the total number of trials.
\[ \begin{equation} y_1, \dots, y_J \sim \mathrm{multinomial}(p_1, \dots, p_J, n) \end{equation} \tag{12.1}\]
We can begin by thinking about the case where n=1. In this case our \(y\) vector will equal 1 for one category and 0 for all other categories. Actually, data like this could be represented using a categorical distribution, which is just like the multinomial distribution except the n is always equal to 1. The multinomial distribution is to the categorical distribution as the binomial distribution is to the Bernoulli distribution (see Chapter 10). We will focus on the multinomial distribution here, however, we will provide an example of how to fit our model using a categorical distribution a bit later in the chapter.
Below we draw 10 instances of a four-category multinomial variable with probabilities of 0.4, 0.1, 0.2, and 0.3 for the first, second, third and fourth categories respectively. In each case, the variable we sample has n=1.
Each individual observation (column) consists of only a single one and three zeros. However, in the code below we can see that if we average across a large number of such observations (across rows), our estimates of \(p_1, \dots, p_J\) closely match the real parameters.
rowMeans (rmultinom (10000, 1, c(.4,.1,.2,.3)))
## [1] 0.3918 0.1023 0.2008 0.3051Below we sample ten more multinomial variables, this time each with n=100. We can see that we get a distribution of values across the four outcomes for each variable that resembles the values of \(p\) we defined for each outcome. Note that when we average across these observations, we get a good estimate of the \(p\) parameter associated with each outcome.
set.seed (1)
multinomial_variable = rmultinom (10, 100, c(.4,.1,.2,.3))
multinomial_variable
## [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10]
## [1,] 42 37 41 41 41 37 36 47 31 37
## [2,] 10 15 7 14 8 14 13 10 14 12
## [3,] 24 21 18 15 19 18 22 16 22 22
## [4,] 24 27 34 30 32 31 29 27 33 29
# the row means closely approximate p
rowMeans(multinomial_variable)/100
## [1] 0.390 0.117 0.197 0.296Multinomial regression is just what it sounds like: A regression model for the prediction of multinomial outcome variables. There are many ways to think about multinomial regression. We’re going to present one that’s consistent with the way we presented logistic regression in Chapter 10 and the way we’ve been presenting regression models in general in this book. For a more complete treatment of the topic please see Agresti (2003), Agresti (2018), and Kruschke (2014). Before beginning the explanation we can ‘spoil’ the ending: The number of trials (\(n\)) and the number of possible outcomes (\(J\)) are aspects of your data and experimental design, and are known to you before you fit any model. Thus, multinomial logistic regression effectively consists of estimating \(p_j\), or values related to it, for each category as a function of your independent variables. In other words, multinomial models let you predict how probable it is that you will see each of the outcome categories, given your independent variables and model structure.
12.2.1 Multinomial logits and the softmax function
Multinomial regression is a surprisingly ‘simple’ extension of the concepts underlying logistic regression. When we discussed logistic regression in Chapter 10, we introduced the inverse logit function (the link function for logistic regression). In Section 10.4.2), we discussed the fact that this function converts log odds to probabilities by arbitrarily setting the ‘count’ of failures to 1 and modeling only the expected ‘count’ of outcomes (i.e. \(e^z\)). We see in equation Equation 12.2 that the probability that \(y=1\) (i.e. a ‘success’) is equal to the ratio of the expected number of successes over the sum of the total number of outcomes.
\[ \begin{equation} \begin{split} P(y=1) = \frac{\mathrm{success}}{\mathrm{failure}+\mathrm{success}} = \frac{e^{z}}{1+e^{z}} \end{split} \end{equation} \tag{12.2}\]
The equation above works when we have exactly two categories. However, it can be modified so that it can be applied to three or more categories. First, we assume that rather than exactly two possible outcomes, there are now \(J\) outcomes, where \(J\) is some integer larger than one. If we stick to our interpretation of the value of \(e^{z}\) as an expected count for that category, then \(e^{z_j}\) is the expected count for category \(j\). To find the probability of observing category \(j\) we find the ratio of the ‘count’ of \(j\) over the sum of all possible outcomes (i.e. the sum of counts for all categories). This is just an extension of the sum of successes and failures when we had only two categories as in equation Equation 12.2.
\[ \begin{equation} \begin{split} P(Y=j) = \frac{e^{z_j}}{\sum_{j=1}^{J} e^{z_j}} \end{split} \end{equation} \tag{12.3}\]
The function above is called the softmax function, and it is basically a generalization of the inverse logit function to more than two categories. When we did dichotomous logistic regression, we modeled only the ‘count’ of one variable, the one for ‘success’. The ‘count’ for the category set to ‘failure’ was not modeled and was set to 1 (by setting \(z=0\), and \(e^{0}=1\)) for all cases. When we model multinomial data we follow the same convention and set one category as the ‘reference’. To do this, we modify the equation above to pull the first category out of the summation and set its count to 1 for all situations, as in equation Equation 12.4. Notice that the summation in the denominator of the fraction now begins at two. In Chapter 10 we said that you can think of the inverse logit function as relating expected successes relative to one failure. You can think of the softmax function as relating the expected number of outcomes of one category to the sum of the expected number of outcomes of all categories, where one of the counts (the reference category) has been set to one.
\[ \begin{equation} \begin{split} P(Y=j) = \frac{e^{z_j}}{1 + \sum_{j=2}^{J} e^{z_j}} \end{split} \end{equation} \tag{12.4}\]
The equation above results in a set of probabilities which can serve as the multinomial parameters, \(p_1, \dots , p_j\), for categories \(1, \dots, J\). These probabilities are modeled by estimating \(z_j\) as the linear combination of our predictor variables based on some unknown parameters (\(\beta_j\)). Each outcome has a different prediction equation for \(z_j\) so that a multinomial regression has \(J\) prediction equations, one for each outcome. In addition, each prediction equation combines the same \(x_k\) predictors, albeit in a category-specific way (based on the \(\beta_j\) parameters for that category). This can be seen in the equation below representing the prediction of \(z_j\) for category \(j\) based on the coefficients (but not predictors) specific to that outcome.
\[ \begin{equation} z_j = \beta_j + \beta_{j1} \cdot x_1 + \beta_{j2} \cdot x_2 + \dots + \beta_{jk} \cdot x_k \end{equation} \tag{12.5}\]
The prediction equation for the ‘reference’ category (brm uses the alphabetically-first category) is fixed to have a value of 0 for \(z_j\). One way to accomplish this is to not estimate parameters for the reference category and instead fix these to zero, as seen in Equation 12.6.
\[ \begin{equation} \begin{split} z_1 = \alpha_1 + \beta_{11} \cdot x_1 + \beta_{12} \cdot x_2 + \dots + \beta_{1k} \cdot x_k \\ z_1 = 0 + 0 \cdot x_1 + 0 \cdot x_2 + \dots + 0 \cdot x_k \\ z_1 = 0 \end{split} \end{equation} \tag{12.6}\]
When we carried out logistic regression, the variable \(z\) was referred to as a logit. Sometimes the \(z_j\) values involved in multinomial regression are called multinomial logits to highlight their similarity to the dichotomous logit. More often, these values are referred to as the score for each category. The ‘meaning’ of the score is somewhat vaguely defined, in part because there are many ways to think about it. One way to think of the score is that it is a latent variable associated with each category that relates to the probability of observing that category given the values of the independent variables.
Latent variables are variables that are inferred mathematically using some statistical model, but are not observed directly. For example, in our logistic regression model in Chapter 10 we predicted the perception of femaleness. We did this by predicting variation in the logit of the probability of observing a female response as a function of speaker vocal-tract length. The logit of this probability can be thought of as a latent variable representing something like ‘femininity/femaleness’ in the mind of the listener. Based on the ‘feeling’ of voice femininity that the listener has, they produce the surface variable that we do measure: A binary classification of a speaker as female/male. In this view the realization of the response variable (a female classification) is the result of a secret, latent, variable that is not directly observed (or observable) by us, but which we assume underlies the process.
In our four-category multinomial model we can imagine four latent variables, the score for each category. We can think of these as the ‘feeling’ the listener has regarding the ‘boyness’, ‘girlness’, ‘manness’, and ‘womanness’ of the voice. What is the ‘boyness’ of a voice? The difficult-to-quantify internal knowledge a listener has that the speaker is a boy, as opposed to some other sort of speaker. These latent variables are not directly measured in our experiment but we think they underlie the classification of speakers into categories. Thus, by observing the categorization of speakers, we can try to make inferences about these underlying variables, and the way that they relate to our predictors.
12.2.2 Comparison to logistic regression
At this point we’ve laid out the basics of multinomial regression, however, our presentation has been very quick and fairly abstract. To make our example more concrete we’re going to talk about a small toy example using a single predictor and three categories, and show that a multinomial model with a single quantitative predictor is a modest extension of a logistic regression model with a single quantitative predictor.
Let’s assume that we’re interested in the prediction of a variable \(Y\) with two outcome categories, 1 and 2. We arbitrarily set 2 to ‘success’ and make it equal to 1 in the vector representing the dependent variable. Supposed we are interested in predicting the probability of observing a success, i.e. \(Y=2\), using a single quantitative predictor \(x\). We model the logit of the probability of observing a success using a line with a given intercept and slope for the quantitative predictor. We set the value of failures to 0 logits for all cases, which is equivalent to representing the failure category with a horizontal line whose intercept is zero. So, our logistic model can be thought of as consisting of the two prediction equations in Equation 12.7, presented in the top left plot of Figure 12.1.
\[ \begin{equation} \begin{split} z_1= 0 + x \cdot 0 \\ z_2= -3 + x \cdot -3 \end{split} \end{equation} \tag{12.7}\]
show plot code
x = seq (-3,3,0.1)
z1 = 0 + x*0
z2 = -3 + x*-3
z3 =-3 + x*3
scores = cbind (z1,z2,z3)
softmax = function (scores){
ps = scores*0
for (i in 1:nrow (scores)){
ps[i,] = exp(scores[i,]) / sum(exp(scores[i,]))
}
ps
}
ps = softmax (scores)
ps2 = softmax (scores[,-3])
par (mfrow = c(2,2), mar=c(.2,4,.2,1), oma = c(4,1,.5,0))
plot (x, z2, type = 'l',col=4, ylim = c(-15,8),xlim=c(-3,3),xaxt='n',
ylab = "Logit",lwd=3,cex.lab=1.3,xaxs='i')
grid()
lines (x,z1,col=1,lwd=3)
plot (x, ps2[,2], type = 'l',col=4, ylim = c(0,1),xlim=c(-3,3),xaxt='n',
ylab = "P(Y=C)",lwd=3,cex.lab=1.3,xaxs='i')
grid()
lines (x,ps2[,1],col=1, lty=2,lwd=3)
plot (x, z2, type = 'l',col=4, ylim = c(-15,8),xlim=c(-3,3),
ylab = "Score",lwd=3,cex.lab=1.3,xaxs='i')
grid()
lines (x,z1,col=1,lwd=3)
lines (x,z3,col=2,lwd=3)
mtext (side=1, "Predictor", line=2.7,lwd=3)
plot (x, ps[,2], type = 'l',col=4, ylim = c(0,1),xlim=c(-3,3),
ylab = "P(Y=C)",lwd=3,cex.lab=1.3,xaxs='i')
grid()
lines (x,ps[,1],col=1,lwd=3)
lines (x,ps[,3], col=2,lwd=3)
mtext (side=1, "Predictor", line =2.7)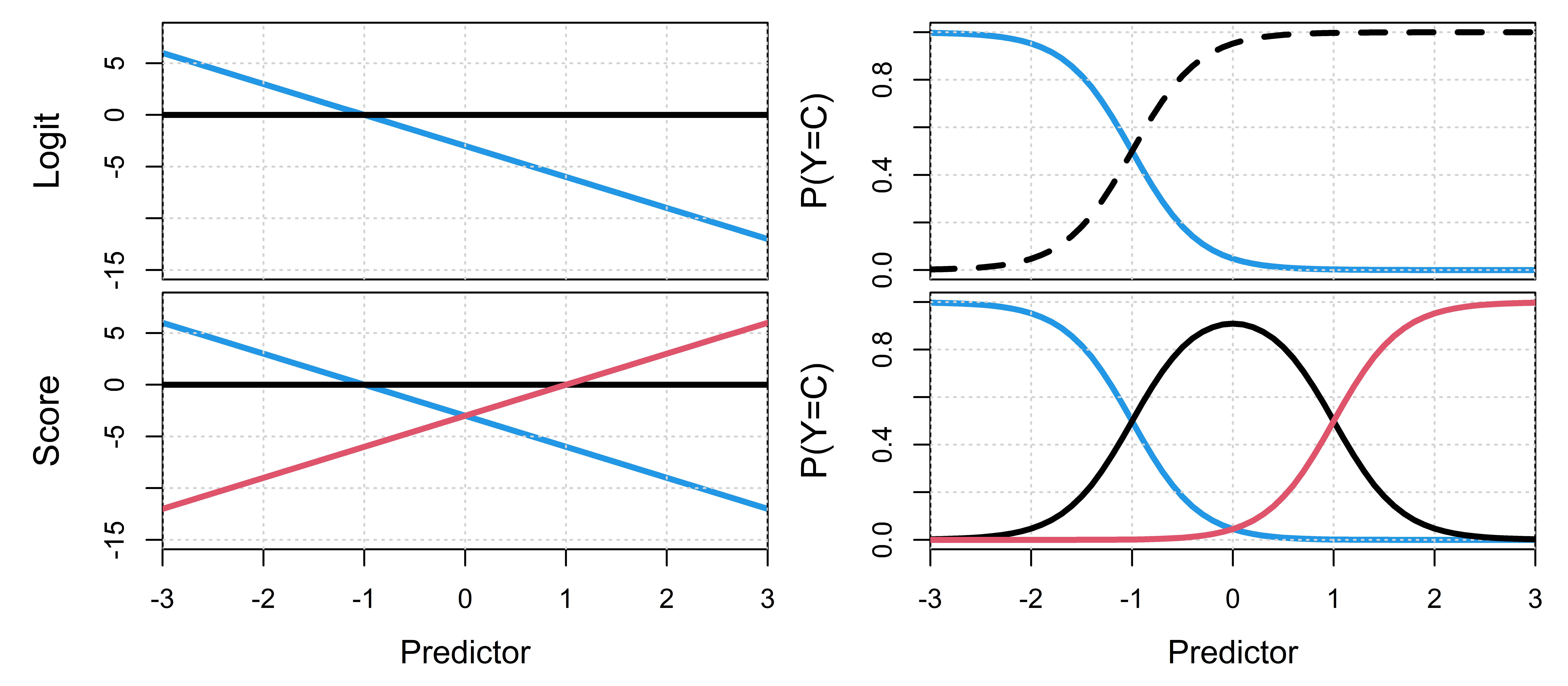
Rather than use the inverse logit function to convert logits to probabilities, we will use the softmax function (introduced above), since we know that these are equivalent when there are two categories. Below, we see that when we set the ‘count’ for failures to 1, i.e. set \(z_1=0\) so that \(e^{z_1}=1\), the softmax function really does ‘become’ the inverse logit function when calculating the probability of a success (\(P(Y=2)\)). We also use this approach to calculate the probability of a failure (\(P(Y=1)\)). Of course, this is not usually done because \(P(Y=2) = 1-P(Y=2)\), however, we do this explicitly here for illustrative purposes.
\[ \begin{equation} \begin{split} P(Y=2) = \frac{e^{z_2}}{e^{z_1} + e^{z_2}} = \frac{e^{z_2}}{1 + e^{z_2}} \\ \\ P(Y=1) = \frac{e^{z_1}}{e^{z_1} + e^{z_2}} = \frac{1}{1 + e^{z_2}} \end{split} \end{equation} \tag{12.8}\]
You can use the code below to recreate the lines in the top right row of Figure 12.1, and to find the probabilities in the top right plot. The probability of observing a success (category 2, blue line) looks just like the sigmoid curves we saw when doing a logistic regression in Chapter 10. In Figure 12.1 we also plot the curve associated with failures. Normally this is not plotted for logistic regression since it is just the curve for successes mirrored about the line at p=0.5.
# predictor from -3 to 3
x = seq (-3,3,0.1)
# lines for failures
z_1 = 0 + x*0
# line for successes
z_2 = -3 + x*-3
scores = cbind (z1,z2)
# softmax function, exponentiate and divide by sum
probabilities = exp (scores) / rowSums (exp (scores))
# simple version of the top right plot in Figure 12.1
plot (probabilities[,1], type = 'l')
lines (probabilities[,2], type = 'l')To turn our logistic regression into a multinomial regression, let’s imagine we want to model a third outcome category, \(Y=3\). We add a line predicting the ‘logit’, now a score, for this category in Equation 12.9. We now have three lines predicting scores (bottom left, Figure 12.1), one of which is still horizontal (\(z_1\)). This is no longer a ‘failure’ but rather the reference category.
\[ \begin{equation} \begin{split} z_1= 0 + x \cdot 0 \\ z_2= -3 + x \cdot -3 \\ z_3= -3 + x \cdot 3 \end{split} \end{equation} \tag{12.9}\]
To get the predicted probability of observing each outcome we enter these scores into the softmax function, in ‘expanded’ form in Equation 12.10.
\[ \begin{equation} \begin{split} P(Y=3) = \frac{e^{z_3}}{1 + e^{z_2} + e^{z_3}} \\ \\ P(Y=2) = \frac{e^{z_2}}{1 + e^{z_2} + e^{z_3}} \\ \\ P(Y=1) = \frac{1}{1 + e^{z_2} + e^{z_3}} \end{split} \end{equation} \tag{12.10}\]
You can use the code below to recreate this process, and the lines in the bottom row of Figure 12.1. Note that all we’ve done is add a third line and a third score. Apart from that, the process is the same as it was for logistic regression.
# predictor from -3 to 3
x = seq (-3,3,0.1)
# lines for reference category
z_1 = 0 + x*0
# line for category two
z_2 = -3 + x*-3
# line for category three
z_2 = -3 + x*3
scores = cbind (z1,z2,z3)
# softmax function, exponentiate and divide by sum
probabilities = exp (scores) / rowSums (exp (scores))
# simple version of the top right plot in Figure 12.1
plot (probabilities[,1], type = 'l',ylim=c(0,1))
lines (probabilities[,2], type = 'l')
lines (probabilities[,3], type = 'l')There is one important difference between logistic and multinomial regression related to classification, or expected outcomes, based on probabilities and scores. When there are only two categories, a logit greater than 0 implies a probability greater than 0.5 of success. In previous chapters, we used this characteristic to talk about expected outcomes in specific parts of the stimulus space. Basically, in any part of the stimulus space where a positive logit is predicted, you expect to observe a ‘success’ (1). In any part of the stimulus space where a negative logit is predicted you expect to observe a ‘failure’ (0). Since the logit of failures is always fixed at exactly zero, another way to look at this is that the category with the greatest predicted logit value is expected.
When there are more than two possible outcome categories, there is no guarantee that a positive score for a given outcome means that category is expected. However, it is still the case that the category with the greatest score in a given condition is the predicted outcome for that condition. Another thing to keep in mind is that, unlike in logistic regression, the most probable outcome may not be very probable in a multinomial model. For example, if there are five outcome categories the most probable predicted outcome could have a probability of only 0.24 if the other categories had probabilities of 0.19. Thus, we see that in multinomial regression the most probable outcome doesn’t have to be very probable, it just needs to be more probable than the alternatives.
12.2.3 Data and research questions
We load our packages and data below. We also add a new variable, \(y\), representing our multinomial outcome. This variable represents observations of a vector of length four whose first, second, third, and fourth elements represent observed outcomes of ‘boy’, ‘girl’, ‘man’, and ‘woman’, respectively. After this we add a variable called size that always equals 1 because each row in our data frame represents the observation of a single outcome. Of course, the size information may be higher than 1 for your data if some of your rows represent trials where n>1. Finally, we process our quantitative predictors in the same way as in the previous chapters.
library (brms)
library (bmmb)
data (exp_data)
# new dependent variable
exp_data$y = cbind(b = as.numeric(exp_data$C=='b'),
g = as.numeric(exp_data$C=='g'),
m = as.numeric(exp_data$C=='m'),
w = as.numeric(exp_data$C=='w'))
# variable representing the size (n) of each observation. They are all 1.
exp_data$size = 1
# preparation of quantitative predictors as in previous chapters
exp_data$vtl_original = exp_data$vtl
exp_data$vtl = exp_data$vtl - mean (exp_data$vtl)
exp_data$f0_original = exp_data$f0
exp_data$f0 = exp_data$f0 - mean(exp_data$f0)
exp_data$f0 = exp_data$f0 / 100Below, we print out the first six instances of our dependent variable and compare this to the first six values of the C (apparent category) variable in our data frame. Each row in the matrix below represents a single observation of our multinomial variable. We can see that the dependent variable indicates which category was selected for a given trial using a 1 in the appropriate column and a 0 in the others.
# first 6 elements of dependent variable
head (exp_data$y)
## b g m w
## [1,] 0 1 0 0
## [2,] 0 0 0 1
## [3,] 0 1 0 0
## [4,] 0 1 0 0
## [5,] 1 0 0 0
## [6,] 1 0 0 0
# first 6 elements of apparent speaker category factor
head (exp_data$C)
## [1] "g" "w" "g" "g" "b" "b"We will try to use our data to answer the following research question:
(Q1) Can we use speaker f0 and VTL to predict apparent speaker category?
Keep in mind we’re trying to predict apparent and not veridical speaker category. Our consideration of the accuracy or utility of this model will depend on how well it represents listener classifications of speakers, no matter how wrong or right these may be. Thus, a model with perfect classification of speakers into veridical categories is not the goal: If listeners make predictable mistakes we want the model to make the same ‘mistakes’.
12.2.4 Description of our model
Our model formula is largely similar to those we have seen before, with two minor changes. Beside the dependent variable, separated by a pipe (|), we need to include a variable that indicates the integer number of trials for each observation. For us this will always be 1, but in many situations this can be a wider range of values. We want to predict our counts of categorizations with respect to the value of speaker VTL and f0, and so the model formula we are going to use is seen below.
y|trials(size) ~ vtl+f0 + (vtl+f0|x|L) + (1|y|S)
Since we’re predicting category membership with two quantitative predictors, we know that the surfaces our model defines are planes. In the models we’ve fit to this point, we had a single dependent variable and a single plane for a single condition. So, our model formulas previously were something like:
y ~ vtl+f0 + (vtl+f0|S) + (1|L)
However, in a multinomial regression we have one plane for each response category, so our formula above could really be thought of as:
y_1 ~ vtl+f0 + (vtl+f0|x|L) + (1|y|S)
y_2 ~ vtl+f0 + (vtl+f0|x|L) + (1|y|S)
y_3 ~ vtl+f0 + (vtl+f0|x|L) + (1|y|S)
y_4 ~ vtl+f0 + (vtl+f0|x|L) + (1|y|S)
As noted in Section 8.6.1, the |x| notation tells our model that the L predictors are related across the prediction equations for each outcome category. In doing this, we estimate the listener effects as coming from a single nine-dimensional normal distribution rather than three separate three dimensional normal variables. Similarly, by adding |y| before our S predictor we model the speaker effects for each outcome category as coming from a single three-dimensional normal distribution rather than three unidimensional ones, and also model the correlations between the dimensions.
Since our multinomial model predicts a response surface for each outcome category, our multinomial model will involve four times as many coefficients as an equivalent Gaussian model. Of course, we noted above that the coefficients of the reference category are set to zero and not estimated. This means that, in general, a multinomial model predicting a variable with \(J\) possible outcomes results in \(J-1\) times as many parameters as an equivalent model predicting a quantitative variable. Our full model specification is pretty large, so we split it into two parts. First, the part specifying our predictions and combinations of predictors:
\[ \begin{equation} \begin{split} y_{1[i]},y_{2[i]},y_{3[i]},y_{4[i]} \sim \mathrm{multinomial}(p_{1[i]},p_{2[i]},p_{3[i]},p_{4[i]}, size_{[i]}) \\ \\ \mathrm{for} \; j = 1, \dots, 4\\ \\ p_{j[i]} = \frac{e^{z_{j[i]}}}{\sum_{j=1}^{J} e^{z_{j[i]}}} \\ \\ z_{j[i]} = \mathrm{a}_j + b_{j[i]} \cdot \mathrm{vtl}_{[i]} + c_{j[i]} \cdot \mathrm{f0}_{[i]} \\ a_{j[i]} = \mathrm{Intercept}_j + L_{j[\mathsf{L}_{[i]}]} + S_{j[\mathsf{S}_{[i]}]} \\ b_{j[i]} = VTL_j + VTL_j \colon L_{j[\mathsf{L}_{[i]}]} \\ c_{j[i]} = F0_j + F0_j \colon L_{j[\mathsf{L}_{[i]}]} \\ \end{split} \end{equation} \tag{12.11}\]
And a plain English description of this part:
Our vector of counts (\(y_{1[i]},y_{2[i]},y_{3[i]},y_{4[i]}\)) is assumed to come from a multinomial distribution with unknown parameters \(p_{1[i]},p_{2[i]},p_{3[i]},p_{4[i]}\), and an n equal to 1 (size) for all trials. For each of our j response categories, the probability of observing that outcome on a trial i, \(p_{j[i]}\), is found by combining the score for each category for that trial (\(z_{j[i]}\)) using the softmax link function. The score for a trial is equal to a trial-dependent combination of an intercept, an effect for VTL, and an effect for f0. The intercept for category j for trial i (\(a_{j[i]}\)) varies according to an overall model intercept for that category (\(\mathrm{Intercept}_{j}\)) and speaker (\(S_{j[\mathsf{S}_{[i]}]}\)) and listener-dependent (\(L_{j[\mathsf{L}_{[i]}]}\)) variations from this. The VTL slope for category j for trial i (\(b_{j[i]}\)) varies according to an overall VTL slope for that category (\(VTL_{j}\)) and listener-dependent (\(VTL \colon L_{j[\mathsf{L}_{[i]}]}\)) variations from this. The f0 slope for category j for trial i (\(c_{j[i]}\)) varies according to an overall f0 slope for that category (\(F0_{j}\)) and listener-dependent (\(F0 \colon L_{j[\mathsf{L}_{[i]}]}\)) variations from this.
In Equation 12.12 we specify the rest of our model (the priors):
\[ \begin{equation} \begin{split} \mathrm{for} \; j = 2, \dots, 4\\ \mathrm{Intercept}_j \sim \mathrm{t}(3, 0, 3) \\ VTL_j, F0_j \sim \mathrm{t}(3, 0, 3) \\ \sigma_{L_j}, \sigma_{VTL_j \colon L_j}, \sigma_{F0_j \colon L_j}, \sigma_{S_j} \sim \mathrm{t}(3, 0, 3) \\ \\ \begin{bmatrix} S_{2[\bullet]} \\ S_{3[\bullet]} \\ S_{4[\bullet]} \end{bmatrix} \sim \mathrm{MVNormal} \left(\, \begin{bmatrix} 0\\ 0 \\ 0 \\ \end{bmatrix}, \mathrm{\Sigma_S} \right) \\ \\ \begin{bmatrix} L_{2[\bullet]} \\ VTL_2 \colon L_{2[\bullet]} \\F0_2 \colon L_{2[\bullet]} \\ L_{3[\bullet]} \\ VTL_3 \colon L_{3[\bullet]} \\F0_3 \colon L_{3[\bullet]} \\ L_{4[\bullet]} \\ VTL_4 \colon L_{4[\bullet]} \\F0_4 \colon L_{4[\bullet]} \\ \end{bmatrix} \sim \mathrm{MVNormal} \left(\, \begin{bmatrix} 0\\ 0 \\ 0 \\ 0\\ 0 \\ 0 \\ 0\\ 0 \\ 0 \\\end{bmatrix}, \mathrm{\Sigma_L} \right) \\ R_S \sim \mathrm{LKJCorr} (2) \\ R_L \sim \mathrm{LKJCorr} (2) \end{split} \end{equation} \tag{12.12}\]
And provide a plan English description of this second part:
We specify priors for response categories 2 to four since all parameters are set to zero for the first (reference) category. The ‘fixed’ effects and correlations were given prior distributions appropriate for their expected range of values. The three speaker dependent terms for each listener (one for each modeled outcome category) were drawn from a three dimensional normal distribution with standard deviations (\(\sigma_{S_2}, \sigma_{S_2}, \sigma_{S_3}\)) and a correlation matrix (\(R_S\)) that was estimated from the data. The nine listener effects were drawn from a nine-dimensional normal distribution with standard deviations (\(\sigma_{L_2}, \sigma_{VTL_2 \colon L_2}, \sigma_{F0_3 \colon L_3}, \, \dots\)) and a correlation matrix (\(R_L\)) that was estimated from the data.
12.2.5 Fitting the model
There is one major difference in how we need to specify priors for multinomial models: We need to specify priors individually for each response category with modeled parameters. This is done by passing the name of the categorical variable to the dpar parameter. Above, we named our response variables according to the letters we have been using throughout this text. We can see this below.
colnames (exp_data$y)
## [1] "b" "g" "m" "w"The name passed to dpar will be muCategory where Category corresponds to the category name. This means we need to specify priors for mug, mum, and muw, but not mub. We specify our priors below:
multinomial_prior =
c(set_prior("student_t(3, 0, 3)", class = "Intercept",dpar="mug"),
set_prior("student_t(3, 0, 3)", class = "b",dpar="mug"),
set_prior("student_t(3, 0, 3)", class = "sd",dpar="mug"),
set_prior("student_t(3, 0, 3)", class = "Intercept",dpar="mum"),
set_prior("student_t(3, 0, 3)", class = "b",dpar="mum"),
set_prior("student_t(3, 0, 3)", class = "sd",dpar="mum"),
set_prior("student_t(3, 0, 3)", class = "Intercept",dpar="muw"),
set_prior("student_t(3, 0, 3)", class = "b",dpar="muw"),
set_prior("student_t(3, 0, 3)", class = "sd",dpar="muw"),
set_prior("lkj_corr_cholesky (2)", class = "cor"))And here is the code to fit our model, using the multinomial family for the first time:
# Fit the model yourself
model_multinomial =
brms::brm (y|trials(size) ~ vtl+f0 + (vtl+f0|x|L) + (1|y|S),
data=exp_data, family="multinomial", chains=4, cores=4,
warmup=1000, iter = 5000, thin = 4, prior = multinomial_prior)# Or download it from the GitHub page:
model_multinomial = bmmb::get_model ("12_model_multinomial.RDS")By the way, if you wanted to fit the model above using the categorical distribution, you would use the code below. Notice that there are only three changes compared to the code to fit our multinomial model above. First, the family parameter is now equal to categorical. Second, the trials specification is now omitted since the number of observations for each trial is now 1. Finally, we can use our vector identifying the outcome category directly as our dependent variable.
model_categorical =
brms::brm (C ~ vtl+f0 + (vtl+f0|L) + (1|S),
data=exp_data, family="categorical", chains=4, cores=4,
warmup=1000, iter = 5000, thin = 4, prior = multinomial_prior)Why would we ever use a multinomial analysis when the categorical distribution seems simpler? First, in some cases individual trials might contain multiple observations such that an individual analysis is not possible. However, a multinomial analysis can also be much faster by substantially reducing the number of individual observations that need to be taken into account when calculating likelihoods. For example, imagine we were only interested in predicting the probability that a listener would report each speaker category. If we run a model like this:
brms::brm (C ~ 1+(1|L), family="categorical", data = exp_data)Then our data has 2085 rows because we have 139 rows for each of 15 listeners. However, if we create a table like this:
table (exp_data$C, exp_data$L)
##
## 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15
## b 18 29 29 40 27 30 41 26 22 5 33 19 51 42 29
## g 38 34 21 39 32 27 23 23 31 45 20 31 12 20 30
## m 39 41 45 40 42 44 44 41 43 43 42 45 42 41 43
## w 44 35 44 20 38 38 31 49 43 46 44 44 34 36 37And use this as our data, then we can treat each column above as a single observation from a multinomial distribution with an n equal to 139. When treated this way, our data has only 15 rows and, as a result, fitting the model below will be much (much) faster.
new_data = data.frame (size = 139, L = factor(1:15))
new_data$y = as.matrix(table (exp_data$L, exp_data$C))
brms::brm (y|trials(size) ~ 1+(1|L), family="multinomial", data = new_data)This is not an option for us since we want to predict categorizations based on the specific combination of listener and speaker, and we only have one observation for each unique combination in our data. However, it is worth considering if a categorical or multinomial analysis might best suit your specific situation.
12.2.6 Interpreting the model
Below we see the model fixed effects, the intercept, VTL slope, and the f0 slope for each category with estimated parameters (i.e. no reference category b). The set of parameters for each category defines a category-specific plane whose value along the \(z\) dimension can be predicted based on the values of f0 and VTL. We have four planes and four values of \(z_j\) for any given location in the two dimensional space defined by f0 and VTL. For each point, we can select as the most probable category the one whose value of \(z_j\) is highest at the point in the space. Another way to look at this is that we select as the most probable outcome the category whose plane is highest at any given location in the space.
brms::fixef (model_multinomial)
## Estimate Est.Error Q2.5 Q97.5
## mug_Intercept -2.4516 0.4508 -3.3802 -1.6018
## mum_Intercept -2.1677 0.4671 -3.1804 -1.3566
## muw_Intercept -0.6317 0.4336 -1.5063 0.2166
## mug_vtl -2.0876 0.3746 -2.8648 -1.3766
## mug_f0 1.4335 0.6987 0.1241 2.8640
## mum_vtl 3.0820 0.5938 2.0761 4.3871
## mum_f0 -2.6146 1.0238 -4.7816 -0.7190
## muw_vtl 0.9464 0.4613 0.0266 1.8643
## muw_f0 2.2001 1.0943 0.1476 4.4024Since the predicted value, the linear predictor, is the score for each category, the parameters above are difficult to interpret in isolation. This is because scores need to be interpreted relative to the baseline category value of 0 (for boys), or relative to one another, rather than absolutely. For example, the negative coefficient for VTL for ‘girl’ (mug_vtl) indicates that a longer VTL made a girl response less likely. We can also see that increasing f0 made a ‘woman’ response more likely (muw_f0), and a ‘man’ response less likely (mum_f0). However, it’s important to remember that these effects are all relative to the effects for the boy category, and not ‘absolute’. For example, the ‘boy’ slope for VTL is 0, meaning it is 2.09 higher than the ‘girl’ slope for the same predictor (which is -2.09). We could say this means that longer VTLs suggest a ‘boy’ response is more likely with respect to a ‘girl’ response. However, a slope of 0 is smaller than the VTL slope for ‘man’ of 3.08. So, a longer VTL means that a ‘boy’ response is less likely with respect to a ‘man’ response. As with all of the parameters in our model, these comparisons can be made more formally with the hypothesis (or short_hypothesis) function as seen below.
# difference in VTL slopes between:
short_hypothesis (model_multinomial,
c("mug_vtl - mum_vtl = 0", # girls and men
"mug_vtl - 0 = 0", # girls and boys
"mum_vtl - 0 = 0")) # men and boys
## Estimate Est.Error Q2.5 Q97.5 hypothesis
## H1 -5.170 0.6927 -6.612 -3.883 (mug_vtl-mum_vtl) = 0
## H2 -2.088 0.3746 -2.865 -1.377 (mug_vtl-0) = 0
## H3 3.082 0.5938 2.076 4.387 (mum_vtl-0) = 0We might wonder how well our model can predict listener judgements. We can do this by finding the predicted probability for each category predicted by our model, as seen below. Note that we use the fitted rather than predict function. This is because we are interested in the expected values predicted by our model. In contrast, predict would give us posterior predictions of the actual dependent variable which would incorporate our modeled noise.
multi_pred_re = fitted (model_multinomial)The result of this is a three dimensional matrix. The first two dimensions represent the usual summary matrices generated by brms, and the third dimension represents the different response categories. Below we see the two-dimensional summary matrix for the first response category (boy). The first column is the posterior probability of category membership for each observation, the second is the standard error for each prediction, and the third and fourth represent the 2.5% and 97.5% credible intervals.
head(multi_pred_re[,,1])
## Estimate Est.Error Q2.5 Q97.5
## [1,] 0.2921 0.1244 0.09423 0.5742
## [2,] 0.2104 0.1036 0.05993 0.4504
## [3,] 0.2804 0.1355 0.07635 0.5892
## [4,] 0.4052 0.1483 0.14678 0.7078
## [5,] 0.4385 0.1408 0.18733 0.7212
## [6,] 0.2218 0.1229 0.04973 0.5123Below we see the same information for the second category (girl):
head(multi_pred_re[,,2])
## Estimate Est.Error Q2.5 Q97.5
## [1,] 0.6190 0.1307 0.3396 0.8427
## [2,] 0.6100 0.1347 0.3243 0.8421
## [3,] 0.6624 0.1380 0.3614 0.8848
## [4,] 0.4507 0.1401 0.1879 0.7200
## [5,] 0.5003 0.1337 0.2454 0.7487
## [6,] 0.6879 0.1367 0.3823 0.9070We can also just select the second dimension from each matrix, resulting in a two-dimensional matrix containing information regarding the posterior probability of being classified into each category across columns, for each observation across rows.
head(multi_pred_re[,1,])
## P(Y = b) P(Y = g) P(Y = m) P(Y = w)
## [1,] 0.2921 0.6190 0.0017775 0.08714
## [2,] 0.2104 0.6100 0.0014059 0.17815
## [3,] 0.2804 0.6624 0.0016820 0.05551
## [4,] 0.4052 0.4507 0.0024148 0.14168
## [5,] 0.4385 0.5003 0.0036467 0.05764
## [6,] 0.2218 0.6879 0.0003084 0.09000Since these are probabilities, the sum of each row equals one because only these four outcomes exist. For example, the first row above tells us that, according to our model, the first observation has a 0.29 probability of being identified as a boy, 0.62 probability of being identified as a girl, 0.002 probability of being identified as a man, and a 0.09 probability of being identified as a woman. We can use the code below to find the ‘winning’ category for each observation, that is, the category with the highest posterior probability in each row. We use the resulting column numbers to get category labels.
# find highest posterior probability from each category
predicted = apply (multi_pred_re[,1,],1,which.max)
head (predicted)
## [1] 2 2 2 2 2 2
# use modal category to get a category label
predicted_category = c("b","g","m","w")[predicted]
head (predicted_category)
## [1] "g" "g" "g" "g" "g" "g"Below, we cross-tabulate predicted and observed classifications. Observed speaker classifications vary across rows, and model predictions of these classifications vary across columns. This is a confusion matrix, introduced in Chapter 5. Correct classifications fall on the main diagonal and all other values indicate mistakes. For example, we see that there were 269 correct classifications of boys as boys, and 78 incorrect predictions of boys as girls. A majority of observations fall along the main diagonal indicating that the model was relatively good at predicting listener behavior.
table (observed_category = exp_data$C, predicted_category)
## predicted_category
## observed_category b g m w
## b 269 78 27 67
## g 74 290 0 62
## m 5 1 624 5
## w 30 19 9 525Below we find the probability of observing a correct classification overall, and individually for each category. We see that the model was able to predict listener judgments with a high degree of accuracy overall, but that some categories (man) were easier to predict than others (boy).
# overall correct
mean (predicted_category == exp_data$C)
## [1] 0.8192
# correct predictions by category
tab = xtabs (~ exp_data$C + predicted_category)
diag(tab) / rowSums(tab)
## b g m w
## 0.6100 0.6808 0.9827 0.900512.2.7 Multinomial models and territorial maps
We discussed territorial maps for two categories along two dimensions in Chapter 11 where we predicted the perception of femaleness based on speaker f0 and VTL. As we discussed then, when categories are represented by planes the boundary between categories is defined by the line formed by the intersection of the planes. When we did (dichotomous) logistic regression in Chapter 11, we found the intersection of one plane with a horizontal plane at \(z=0\). In this chapter we still have a plane such that \(z=0\) for all x and y (the reference category plane), but we also have three other planes with possible non-zero slopes along each dimension. The intersection of the planes representing pairs of categories forms the boundary between those two categories. Since we have 6 unique pairings of our four response categories, we will have six boundary lines.
To find the line formed by the intersection of planes, first we define the values of two planes, \(z_1\) and \(z_2\) based on their respective coefficients as in Equation 12.13. We use \(a\), \(b\), and \(c\) for the intercept, x axis, and y axis slope for the first plane and \(d\), \(e\), and \(f\) for the corresponding coefficients for the second plane.
\[ \begin{equation} \begin{split} z_1 = \mathrm{a} + \mathrm{b} \cdot x + \mathrm{c} \cdot y \\ z_2 = \mathrm{d} + \mathrm{e} \cdot x + \mathrm{f} \cdot y \end{split} \end{equation} \tag{12.13}\]
We set \(z_1\) equal to \(z_2\) to find the place where the planes equal (i.e. their intersection) as in Equation 12.14.
\[ \begin{equation} \begin{split} z_1 = z_2 \\ \mathrm{a} + \mathrm{b} \cdot x + \mathrm{c} \cdot y = \mathrm{d} + \mathrm{e} \cdot x + \mathrm{f} \cdot y \end{split} \end{equation} \tag{12.14}\]
In order to turn Equation 12.14 into an equation resembling that of a line (\(y = a + b \cdot x\)), we need to isolate y on the left hand side as seen in Equation 12.15.
\[ \begin{equation} y = \frac {-\mathrm{a} + \mathrm{d}}{\mathrm{c} - \mathrm{f}} + \frac{-\mathrm{b} + \mathrm{e}}{\mathrm{c} - \mathrm{f}} \cdot x \end{equation} \tag{12.15}\]
The equation above is actually much simpler than it looks. Since the parameters (\(a,b,c,d,e,f\)) are just constants, the intercept (\(-a+d/c-f\)) and slope (\(-b+e/c-f\)) simplify to scalars. As mentioned above, we get 6 such line equations, one for the boundary between all possible pairs of each of the four planes. Figure 12.2 presents each of these six boundaries compared to the modal classification for each speaker.
show plot code
params = fixef (model_multinomial)
params = fixef (model_multinomial, summary = FALSE)
params = cbind (colMeans (params[,1:3]),
colMeans (params[,c(4,6,8)]),
colMeans (params[,c(5,7,9)]))
params = rbind (c(0,0,0), params[1:3,])
tab = table (exp_data$S, exp_data$C)
mod_cat = apply (tab, 1,which.max)
par (mfrow = c(2,3), mar = c(0.2,0.2,0.2,0.2), oma = c(4.5,4.5,0.5,0.5))
plot(model_multinomial$data$vtl,model_multinomial$data$f0, ylim = c(-1.25,1.25),
xlim = c(-2.5,3), pch = 16, col = bmmb::cols[2:5][mod_cat],cex=2,
xaxt='n')
abline (find_intersection (params[1,], params[2,]),lwd=2, col = bmmb::cols[7]) # b vs g
text (1.5,1, "boy-girl", cex=1.5)
plot(model_multinomial$data$vtl,model_multinomial$data$f0, ylim = c(-1.25,1.25),
xlim = c(-2.5,3), pch = 16, col = bmmb::cols[2:5][mod_cat],cex=2,
yaxt='n',xaxt='n')
abline (find_intersection (params[1,], params[3,]),lwd=2, col = bmmb::cols[8]) # b vs m
text (1.5,1, "boy-man", cex=1.5)
legend (-2.3,-0.00, legend=c("b","g","m","w"),col=bmmb::cols[2:5],
pch=16,pt.cex=1.75,bty='n', cex=1.20)
plot(model_multinomial$data$vtl,model_multinomial$data$f0, ylim = c(-1.25,1.25),
xlim = c(-2.5,3), pch = 16, col = bmmb::cols[2:5][mod_cat],cex=2,
yaxt='n',xaxt='n')
abline (find_intersection (params[1,], params[4,]),lwd=2, col = bmmb::cols[9]) # b vs w
text (1.5,1, "boy-woman", cex=1.5)
plot(model_multinomial$data$vtl,model_multinomial$data$f0, ylim = c(-1.25,1.25),
xlim = c(-2.5,3), pch = 16, col = bmmb::cols[2:5][mod_cat],cex=2)
abline (find_intersection (params[2,], params[3,]),lwd=2, col = bmmb::cols[10]) # g vs m
text (1.5,1, "girl-man", cex=1.5)
plot(model_multinomial$data$vtl,model_multinomial$data$f0, ylim = c(-1.25,1.25),
xlim = c(-2.5,3), pch = 16, col = bmmb::cols[2:5][mod_cat],cex=2,
yaxt='n')
abline (find_intersection (params[2,], params[4,]),lwd=2, col = bmmb::cols[13]) # g vs w
text (1.5,1, "girl-woman", cex=1.5)
plot(model_multinomial$data$vtl,model_multinomial$data$f0, ylim = c(-1.25,1.25),
xlim = c(-2.5,3), pch = 16, col = bmmb::cols[2:5][mod_cat],cex=2,
yaxt='n')
abline (find_intersection (params[3,], params[4,]),lwd=2, col = bmmb::cols[14]) # m vs w
text (1.5,1, "man-woman", cex=1.5)
mtext (side=1, "Centered vocal tract length (cm)", outer = TRUE, line=3)
mtext (side=2, "Centered f0 / 100 (Hz)", outer = TRUE, line=3)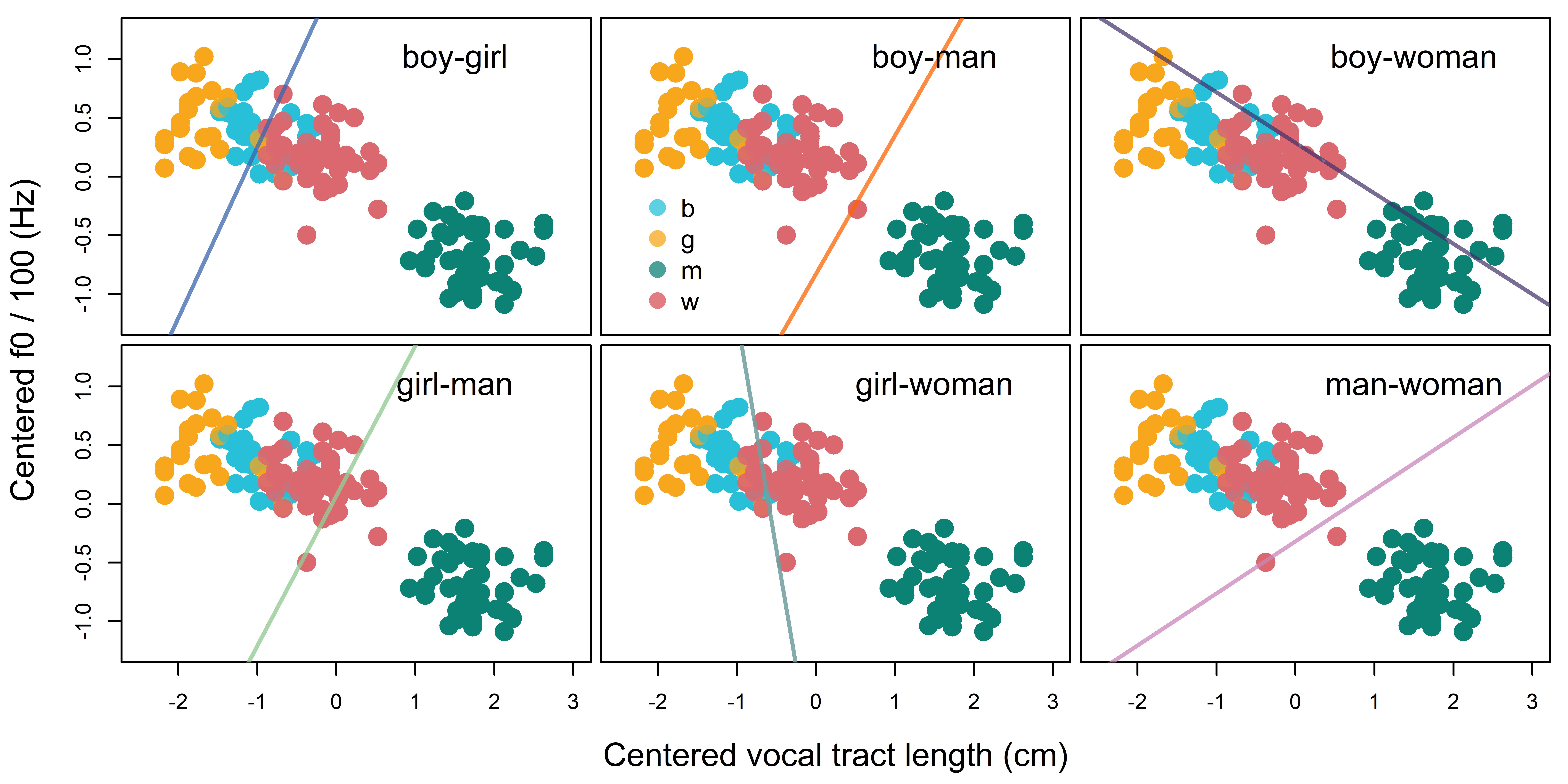
All six boundary lines are presented in the left plot of Figure 12.3. One problem with considering the figure in this way is that many of the boundaries are not very relevant. For example, the boundary between girl and man will hardly matter for classification because the ‘woman’ and ‘boy’ categories fall almost entirely between the ‘girl’ and ‘man’ categories. As a result, listeners will rarely be deciding between a ‘man’ or ‘girl’ response when ‘boy’ and ‘woman’ are available.
The right plot of Figure 12.3 presents only those boundaries that are relevant, resulting in a territorial map of our categories along f0 and VTL (see Section 10.5.5). This can be stated more formally as: The figure contains only those boundaries that represents scores equal to or greater than the maximal score for that location, from among the response categories. For example, in the left plot we see that the boy-woman boundary bisects the area where ‘man’ is a modal response (bottom-right in the left plot). This line segment does not feature in the right plot because it has a lower value than the ‘man’ plane in this area. One way to imagine how territorial maps relate to our planes is to imagine that our planes were opaque but were immaterial such that they can intersect and continue past each other. If we were to look ‘down’ the z axis onto these planes, we would see something very much like the right plot of Figure 12.3; we would see only those plane sections, and only those intersections, that were ‘above’ any other plane.
show plot code
params = fixef (model_multinomial)
params = fixef (model_multinomial, summary = FALSE)
params = cbind (colMeans (params[,1:3]),
colMeans (params[,c(4,6,8)]),
colMeans (params[,c(5,7,9)]))
params = rbind (c(0,0,0), params[1:3,])
tab = table (exp_data$S, exp_data$C)
mod_cat = apply (tab, 1,which.max)
par (mfrow = c(1,2), mar = c(.0,.0,.21,.2),oma=c(4,4,.1,.1))
plot(model_multinomial$data$vtl,model_multinomial$data$f0, ylim = c(-1.25,1.25),
xlim = c(-2.5,3), pch = 16, col = bmmb::cols[2:5][mod_cat])
abline (find_intersection (params[1,], params[2,]),lwd=3, col = bmmb::cols[7])
abline (find_intersection (params[1,], params[3,]),lwd=3, col = bmmb::cols[8])
abline (find_intersection (params[1,], params[4,]),lwd=3, col = bmmb::cols[9])
abline (find_intersection (params[2,], params[3,]),lwd=3, col = bmmb::cols[10])
abline (find_intersection (params[2,], params[4,]),lwd=3, col = bmmb::cols[13])
abline (find_intersection (params[3,], params[4,]),lwd=3, col = bmmb::cols[14])
points(model_multinomial$data$vtl,model_multinomial$data$f0, pch = 16, col = bmmb::cols[2:5][mod_cat], cex = 2)
#points(model_multinomial$data$vtl,model_multinomial$data$f0, pch = 1, col = 1,
# lwd=2, cex=2)
plot(model_multinomial$data$vtl,model_multinomial$data$f0, ylim = c(-1.25,1.25),
xlim = c(-2.5,3), pch = 16, col = bmmb::cols[2:5][mod_cat],ylab="",yaxt="n")
maps = make_map (t(params))
plot_map (maps, colors = adjustcolor (bmmb::cols[2:5], alpha.f=.2), new=FALSE)
points(model_multinomial$data$vtl,model_multinomial$data$f0, pch = 16, col = bmmb::cols[2:5][mod_cat], cex = 2)
#points(model_multinomial$data$vtl,model_multinomial$data$f0, pch = 1, col = 1,
# lwd=2, cex=2)
mtext (side=1, "Centered vocal tract length (cm)", outer = TRUE, line=2.5)
mtext (side=2, "Centered f0 / 100 (Hz)", outer = TRUE, line=2.5)
legend (-0.5,1.3, legend=c("b","g","m","w"),col=bmmb::cols[2:5],
pch=16,pt.cex=1.25,bty='n', cex=1.0, horiz = TRUE)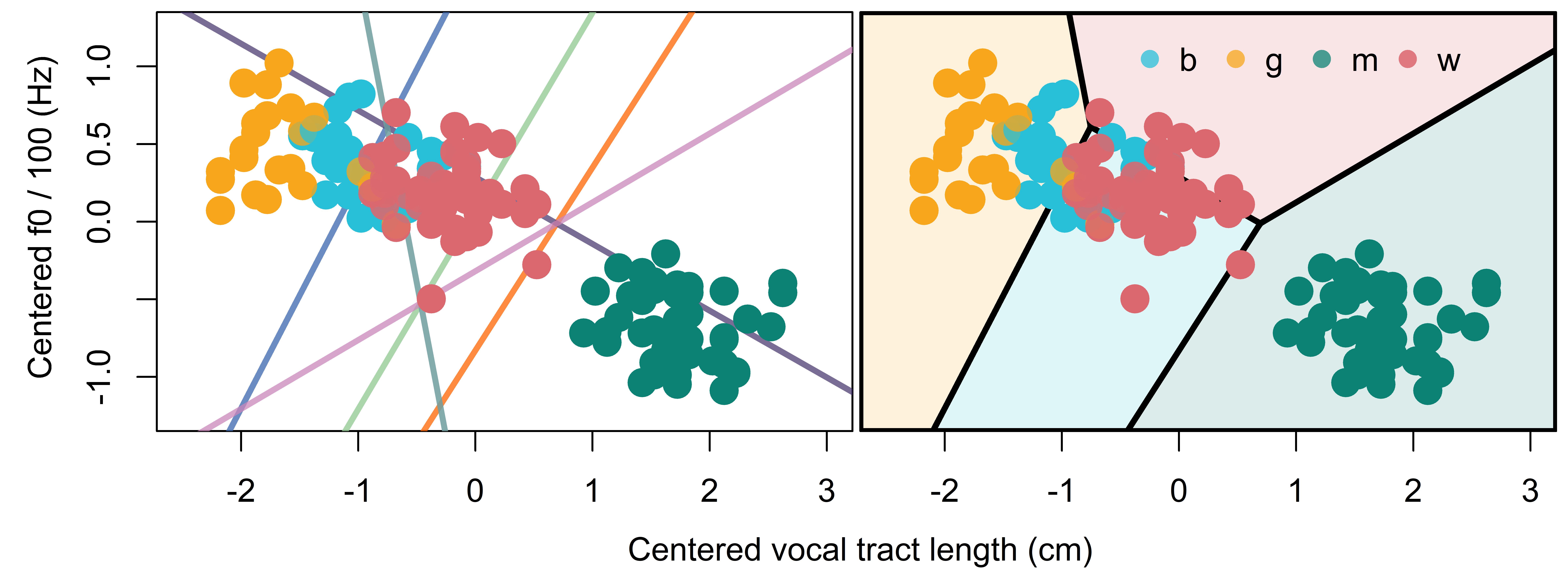
We can find the boundaries of each of the ‘territories’ in the territorial map analytically by finding each boundary, the intersections of all boundaries, and record only the line segments whose scores where greater than or equal to all of the category planes. Each of these steps is generally straightforward but we know of no algorithm to implement this process easily for any number of response categories. The make_map function in the bmmb package can be used to generate territorial maps for any number of response categories in a two-dimensional stimulus space. However, rather than find the territorial map analytically, make_map estimates its characteristics numerically, that is, based on a bunch of guesses and not exact solutions.
The make_map function takes in a matrix representing the coefficients for each of the response categories. In the code below we get estimates for our fixed effects and combine these into a matrix, including a column of zeros to represent the ‘boy’ coefficients. Each column represents the coefficients for a single response category, and intercepts, x axis coefficients and y axis coefficients vary across rows. The rows and columns of the matrix below have been named to make this organization clearer, and you should compare the values below to our model fixed effects shown above (e.g. fixef (model_multinomial)).
parameters = unname (fixef (model_multinomial)[,1])
parameters = cbind(b = c(0, 0, 0),
g = parameters[c(1,4,5)],
m = parameters[c(2,6,7)],
w = parameters[c(3,8,9)])
rownames(parameters)=c("Intercept","vtl_slope","f0_slope")
parameters
## b g m w
## Intercept 0 -2.452 -2.168 -0.6317
## vtl_slope 0 -2.088 3.082 0.9464
## f0_slope 0 1.434 -2.615 2.2001This matrix can be passed to the make_map function, which uses it to calculate the territorial map. It operates in the following manner. First, it generates a 1000 by 1000 point grid spanning the desired x and y axis range. The density of points can be set with the density parameter and the x and y axis range can be set manually or by passing points with the same ranges to the function with the points parameter. After this, the function uses the parameters given to calculate the score for each category, for every point in the grid, and collects the winning category for each location. After this, the convex hull encompassing the points for each category is found, and the polygon representing the convex hull for each category is returned in a list by the function. This approach is not exact, and so is not appropriate for making big claims about your results. However, it can be made arbitrarily accurate by increasing the density of the points, and this approach is sufficient for creating plots to convey the results of your analyses.
territorial_map =
bmmb::make_map (parameters, points = cbind(exp_data$vtl,exp_data$f0))The output of make_map can be passed to the plot_map function to plot the resulting territorial maps easily. We used this process to create the territorial maps of the figures in this chapter.
bmmb::plot_map (territorial_map, colors = bmmb::cols[2:5],xlab="",ylab="")We noticed something very strange when we made the territorial map in Figure 12.3: The ‘boy’ territory contains a large number of woman responses. In addition, the boy-woman boundary seems strangely high along f0. If that is really the boundary between boys and women in the VTL by f0 space, then a large number of adult female speakers fall into territory where ‘boy’ responses are expected. If this is the case then how/why is the model classifying all those women correctly? The answer to this question may lie in Figure 12.4.
In the left plot of Figure 12.4, point colors represent modal listener judgments for each speaker. In the middle plot we see model predictions including speaker and listener random effects. When these are included the model predictions look very much like the listener judgments and the classifications don’t make much sense relative to the territorial map. In the right plot of the same figure we see model predictions when random effects are not included. In this case we see that model predictions do not look like the listener judgments, but the classifications do make sense relative to the territorial map. So, it seems that the random effects are causing this strange behavior in our model.
show plot code
params = fixef (model_multinomial)
params = fixef (model_multinomial, summary = FALSE)
params = cbind (colMeans (params[,1:3]),
colMeans (params[,c(4,6,8)]),
colMeans (params[,c(5,7,9)]))
params = rbind (c(0,0,0), params[1:3,])
tab = table (exp_data$S, exp_data$C)
mod_cat = apply (tab, 1,which.max)
cats = apply(multi_pred[,1,],1,which.max)
#table (exp_data$C, cats)
tab2 = table (exp_data$S, cats)
mod_cat2 = apply (tab2, 1,which.max)
cats = apply(multi_pred_re[,1,],1,which.max)
#table (exp_data$C, cats)
tab2 = table (exp_data$S, cats)
mod_cat3 = apply (tab2, 1,which.max)
par (mfrow = c(1,3), mar = c(.0,.0,.21,.2),oma=c(4,4,2,.1))
plot(model_multinomial$data$vtl,model_multinomial$data$f0, ylim = c(-1.25,1.25),
xlim = c(-2.5,3), pch = 16, col = bmmb::cols[2:5][mod_cat],ylab="")
maps = make_map (t(params))
mtext (side=3, text = "Listener Judgments", line = 0.5)
plot_map (maps, colors = adjustcolor (bmmb::cols[2:5], alpha.f=.2), new=FALSE)
points(model_multinomial$data$vtl,model_multinomial$data$f0, pch = 16, col = bmmb::cols[2:5][mod_cat], cex = 2)
legend (-0.9,1.3, legend=c("b","g","m","w"),col=bmmb::cols[2:5],
pch=16,pt.cex=1.5,bty='n', cex=1.3, horiz = TRUE)
plot(model_multinomial$data$vtl,model_multinomial$data$f0, ylim = c(-1.25,1.25),
xlim = c(-2.5,3), pch = 16, col = bmmb::cols[2:5][mod_cat2],ylab="",yaxt="n")
plot_map (maps, colors = adjustcolor (bmmb::cols[2:5], alpha.f=.2), new=FALSE)
points(model_multinomial$data$vtl,model_multinomial$data$f0, pch = 16, col = bmmb::cols[2:5][mod_cat3], cex = 2)
mtext (side=3, text = "Model Predictions (with RE)", line = 0.5)
plot(model_multinomial$data$vtl,model_multinomial$data$f0, ylim = c(-1.25,1.25),
xlim = c(-2.5,3), pch = 16, col = bmmb::cols[2:5][mod_cat3],ylab="",yaxt="n")
plot_map (maps, colors = adjustcolor (bmmb::cols[2:5], alpha.f=.2), new=FALSE)
points(model_multinomial$data$vtl,model_multinomial$data$f0, pch = 16, col = bmmb::cols[2:5][mod_cat2], cex = 2)
mtext (side=3, text = "Model Predictions (no RE)", line = 0.5)
mtext (side=1, "Centered vocal tract length (cm)", outer = TRUE, line=2.5)
mtext (side=2, "Centered f0 / 100 (Hz)", outer = TRUE, line=2.5)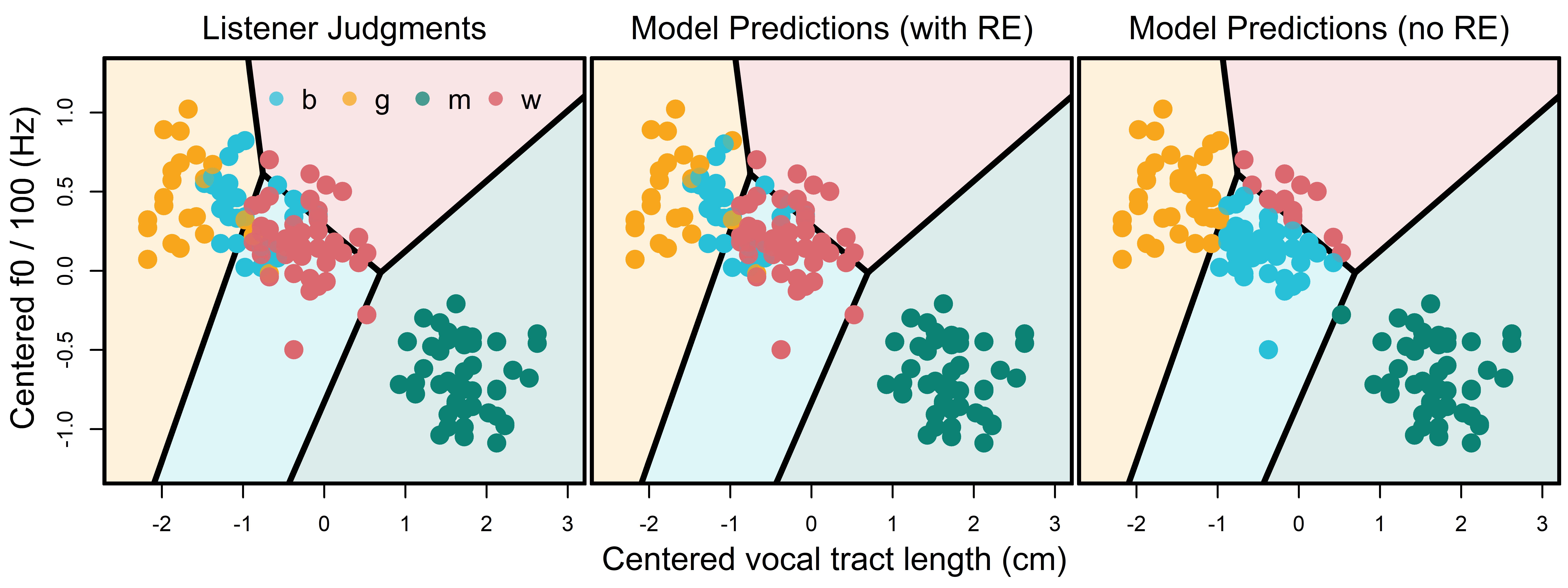
Figure 12.5 presents the speaker random intercepts. Since there are three modeled categories, there are three random intercepts for each speaker. We can see that, in general, the speaker intercepts have small values that vary around zero, and have credible intervals that mostly include zero. However, many female speakers have intercepts that have large non-zero values for the female category, and whose credible intervals omit zero and very small values.
show plot code
sranef = ranef(model_multinomial)$S
tab = table (exp_data$S, exp_data$C_v)
real_cat = apply (tab, 1,which.max)
par (mfrow = c(3,1), mar =c(.1,4,.1,.5), oma = c(.5,0,.5,0))
#brmplot((sranef[,,1]+sranef[,,2]+sranef[,,3])/3, labels = '',
# col = bmmb::cols[real_cat+1], ylim = c(-8,8))
#text (1,7,"Boy category speaker intercepts", pos=4,cex=1.4)
brmplot(sranef[,,1], labels = '', col = bmmb::cols[real_cat+1], ylim = c(-8,8))
text (1,7,"Girl category speaker intercepts", pos=4,cex=1.4)
legend (1,-4.5, legend=c("b","g","m","w"),col=bmmb::cols[2:5],
pch=16,pt.cex=1.5,bty='n', cex=1.1, horiz = TRUE)
brmplot(sranef[,,2], labels = '', col = bmmb::cols[real_cat+1], ylim = c(-8,8))
text (1,7,"Man category speaker intercepts", pos=4,cex=1.4)
brmplot(sranef[,,3], labels = '', col = bmmb::cols[real_cat+1], ylim = c(-8,8))
text (1,7,"Woman category speaker intercepts", pos=4,cex=1.4)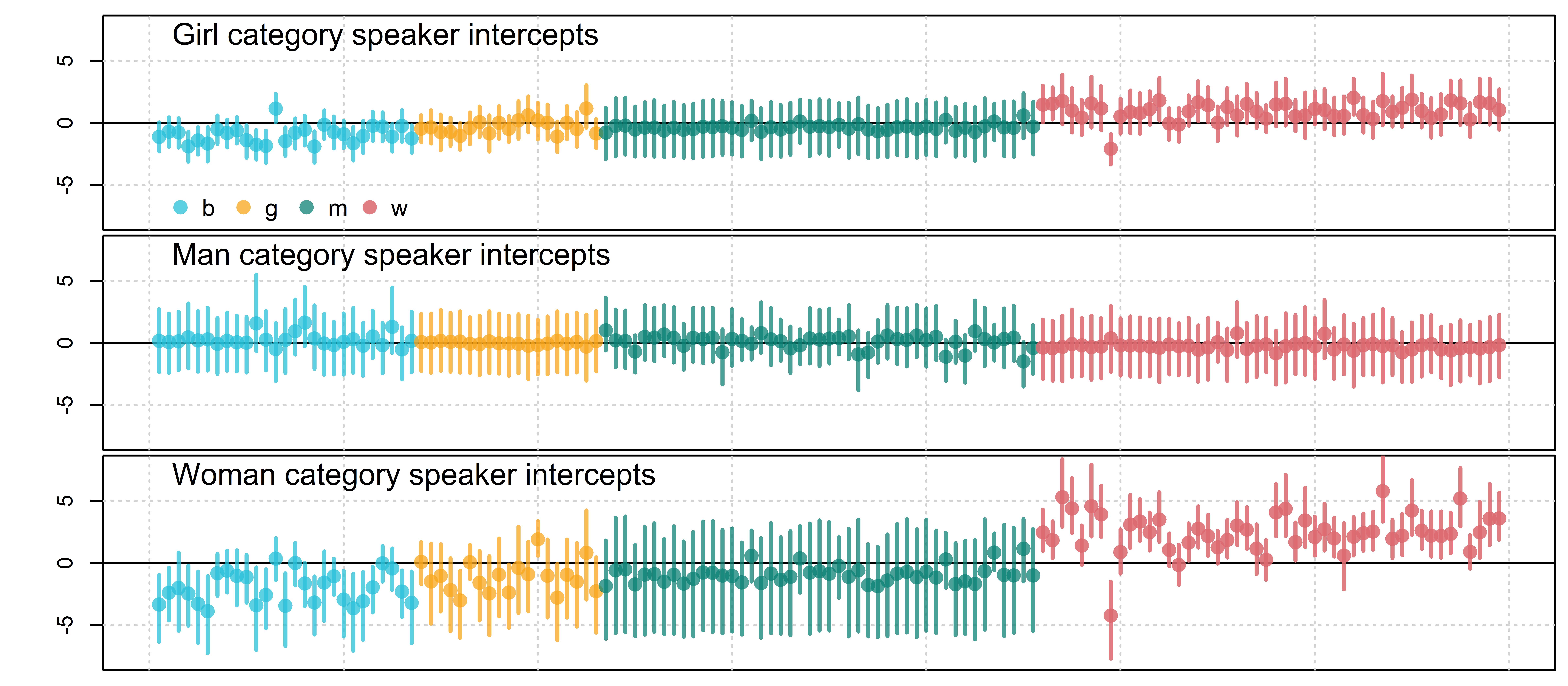
Here’s what we believe is happening. Think of a female response plane with a specific intercept, VTL slope and f0 slope. If female speaker categorizations were very predictable based on the value of this plane at a given location, all the female speaker effects would be zero or near zero. This is because if the score based on the plane explains classification, there is nothing left for the speaker intercept to explain. We can see this sort of behavior for adult male speakers: Adult male speakers are mostly easy to identify because of their unusually low f0s.
In contrast, consider a situation where individual female speakers fall way off the plane, in a seemingly random but consistent manner. For example, imagine a female speaker whose score for the female category is way higher than the plane says it should be, but that every listener seems to behave this way for this speaker. This would mean that this is a consistent classification for this voice that cannot be explained by the score according to the plane. In other words, the classification cannot be predicted based on the linear combination of voice VTL and f0. In such a situation, this consistent classification would be ‘explained’ by the speaker intercept. Effectively, this tells your model “use the general female plane, but move it up/down this much specifically for this speaker”. Why are we moving it up/down for this speaker? Our model can’t tell us, since we’re just modeling this with an intercept (and not an additional predictor). However, it can tell us that the up/down adjustment is consistent for the speaker across listeners and improves model performance.
We can think about what these speaker effects might reflect. The information regarding gender and age in voices is complicated, and likely reflects subtle stylistic and rhythmic cues. Basically, sounding ‘feminine’ and ‘masculine’ is much more complicated than just f0 or VTL (see Section 13.1.6). This is analogous to the fact that women tend to be shorter than men, but it would be ridiculous to suggest that masculinity/femininity are entirely predictable based on the height of a person. Size is perhaps an aspect of perceived femininity/masculinity but the whole of it is substantially more complicated.
We again consider our model prediction accuracy, but this time make predictions without any random effects.
# predictions with no random effects
multi_pred = fitted (model_multinomial, re_formula = NA)# find winners
predicted_no_re = apply (multi_pred[,1,],1,which.max)
# get winner labels
predicted_category_no_re = c("b","g","m","w")[predicted_no_re]
# overall correct
mean (predicted_category_no_re == exp_data$C)
## [1] 0.5871
# correct predictions by category
tab = xtabs (~ exp_data$C + predicted_category_no_re)
diag(tab) / rowSums(tab)
## b g m w
## 0.2993 0.7559 0.9827 0.2504The picture without random effects is a bit grim. Overall correct prediction is not too bad (relative to chance at 25%), however, some of the individual categories are predicted very poorly. As expected based on the territorial maps in Figure 12.4, the accurate prediction of ‘woman’ classifications is particularly bad with only 21% of ‘woman’ responses being accurately predicted as such.
12.2.8 Refitting the model without speaker random effects
The results of our previous model suggest that the inclusion of speaker random effects may lead to some unintended, and undesirable, consequences. The predictions made by our initial model are only good as long as we include the speaker random effects. However, these do not let us understand the prediction of new speakers based solely on speaker f0 and VTL. Further, the territorial map we generate using the model is a bit unreliable because of the influence of the speaker random effects included in the model. We will try fitting the model without speaker effects to see if we get a territorial map that is a better match for our listener judgments. We use the same priors we used before, and fit the same model save for the absence of speaker effects in our formula:
# Fit the model yourself
model_multinomial_noS =
brms::brm (y|trials(size) ~ vtl+f0 + (vtl+f0|x|L), data=exp_data,
family=multinomial(), chains=4, cores=4, warmup=1000,
iter = 5000, thin = 4, prior = multinomial_prior)# Or download it from the GitHub page:
model_multinomial_noS =
bmmb::get_model ("../bmmrmd/models/12_model_multinomial_noS.RDS")We jump straight to plotting the territorial map represented by this model, again comparing listener judgments to model predictions with and without random effects (Figure 12.6). We can see that the territorial maps in the figure seem to be a better match to the data.
show plot code
params = fixef (model_multinomial_noS)
params = fixef (model_multinomial_noS, summary = FALSE)
params = cbind (colMeans (params[,1:3]),
colMeans (params[,c(4,6,8)]),
colMeans (params[,c(5,7,9)]))
params = rbind (c(0,0,0), params[1:3,])
tab = table (exp_data$S, exp_data$C)
mod_cat = apply (tab, 1,which.max)
cats = apply(multi_pred_noS[,1,],1,which.max)
#table (exp_data$C, cats)
tab2 = table (exp_data$S, cats)
mod_cat2 = apply (tab2, 1,which.max)
cats = apply(multi_pred_re_noS[,1,],1,which.max)
#table (exp_data$C, cats)
tab2 = table (exp_data$S, cats)
mod_cat3 = apply (tab2, 1,which.max)
par (mfrow = c(1,3), mar = c(.0,.0,.21,.2),oma=c(4,4,2,.1))
plot(model_multinomial$data$vtl,model_multinomial$data$f0, ylim = c(-1.25,1.25),
xlim = c(-2.5,3), pch = 16, col = bmmb::cols[2:5][mod_cat],ylab="")
maps_noS = make_map (t(params))
mtext (side=3, text = "Listener Judgments", line = 0.5)
legend (-2.5,-0.15, legend=c("b","g","m","w"),col=bmmb::cols[2:5],
pch=16,pt.cex=1.5,bty='n', cex=1.15)
plot_map (maps_noS, colors = adjustcolor (bmmb::cols[2:5], alpha.f=.2), new=FALSE)
points(model_multinomial$data$vtl,model_multinomial$data$f0, pch = 16, col = bmmb::cols[2:5][mod_cat], cex = 2)
plot(model_multinomial$data$vtl,model_multinomial$data$f0, ylim = c(-1.25,1.25),
xlim = c(-2.5,3), pch = 16, col = bmmb::cols[2:5][mod_cat2],ylab="",yaxt="n")
plot_map (maps_noS, colors = adjustcolor (bmmb::cols[2:5], alpha.f=.2), new=FALSE)
points(model_multinomial$data$vtl,model_multinomial$data$f0, pch = 16, col = bmmb::cols[2:5][mod_cat3], cex = 2)
mtext (side=3, text = "Model Predictions (with RE)", line = 0.5)
plot(model_multinomial$data$vtl,model_multinomial$data$f0, ylim = c(-1.25,1.25),
xlim = c(-2.5,3), pch = 16, col = bmmb::cols[2:5][mod_cat3],ylab="",yaxt="n")
plot_map (maps_noS, colors = adjustcolor (bmmb::cols[2:5], alpha.f=.2), new=FALSE)
points(model_multinomial$data$vtl,model_multinomial$data$f0, pch = 16, col = bmmb::cols[2:5][mod_cat2], cex = 2)
mtext (side=3, text = "Model Predictions (no RE)", line = 0.5)
mtext (side=1, "Centered vocal tract length (cm)", outer = TRUE, line=2.5)
mtext (side=2, "Centered f0 / 100 (Hz)", outer = TRUE, line=2.5)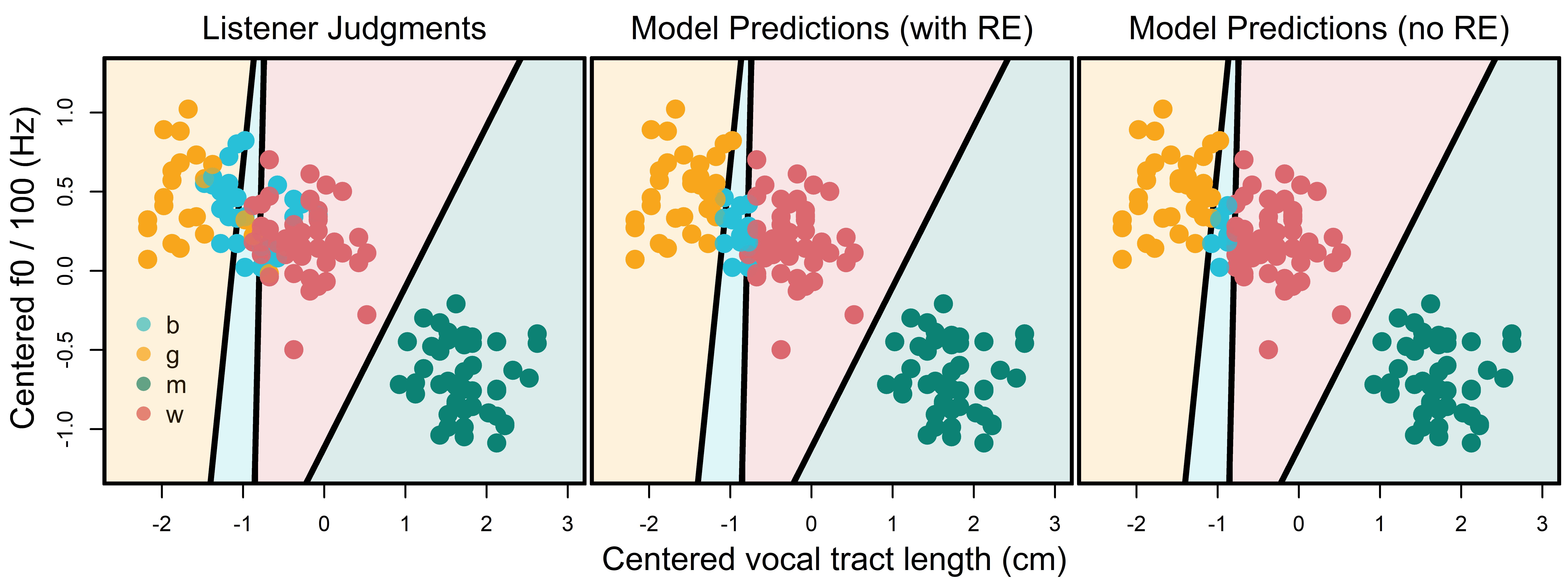
Figure 12.7 compares the territorial maps represented by our models with (left) and without (right) speaker information. In addition to being a better fit to the data, we think the new territorial maps make more ‘sense’. This is because the map with speaker effects suggests that the difference between women and boys was almost entirely due to f0, with women being associated with the higher f0. This is despite the fact that veridical boys, on average, have shorter VTLs and higher f0s than adult females. The new map has basically no effect for f0 between these categories and makes it primarily a VTL difference, which seems to be a better fit to the data.
show plot code
params = fixef (model_multinomial_noS)
params = fixef (model_multinomial_noS, summary = FALSE)
params = cbind (colMeans (params[,1:3]),
colMeans (params[,c(4,6,8)]),
colMeans (params[,c(5,7,9)]))
params = rbind (c(0,0,0), params[1:3,])
tab = table (exp_data$S, exp_data$C)
mod_cat = apply (tab, 1,which.max)
cats = apply(multi_pred_noS[,1,],1,which.max)
tab2 = table (exp_data$S, cats)
mod_cat2 = apply (tab2, 1,which.max)
cats = apply(multi_pred_re_noS[,1,],1,which.max)
tab2 = table (exp_data$S, cats)
mod_cat3 = apply (tab2, 1,which.max)
par (mfrow = c(1,2), mar = c(.0,.0,.0,.2),oma=c(3.5,3.5,2,.1))
plot(model_multinomial$data$vtl,model_multinomial$data$f0, ylim = c(-1.25,1.25),
xlim = c(-2.5,3), pch = 16, col = bmmb::cols[2:5][mod_cat],ylab="")
#maps_noS = make_map (t(params))
mtext (side=3, text = "model_multinomial", line = 0.5)
legend (-0.5,1.3, legend=c("b","g","m","w"),col=bmmb::cols[2:5],
pch=16,pt.cex=1.25,bty='n', cex=1.0, horiz = TRUE)
plot_map (maps, colors = adjustcolor (bmmb::cols[2:5], alpha.f=.2), new=FALSE)
points(model_multinomial$data$vtl,model_multinomial$data$f0, pch = 16, col = bmmb::cols[2:5][mod_cat], cex = 2)
plot(model_multinomial$data$vtl,model_multinomial$data$f0, ylim = c(-1.25,1.25),
xlim = c(-2.5,3), pch = 16, col = bmmb::cols[2:5][mod_cat2],ylab="",yaxt="n")
plot_map (maps_noS, colors = adjustcolor (bmmb::cols[2:5], alpha.f=.2), new=FALSE)
points(model_multinomial$data$vtl,model_multinomial$data$f0, pch = 16, col = bmmb::cols[2:5][mod_cat], cex = 2)
mtext (side=3, text = "model_multinomial_noS", line = 0.5)
mtext (side=1, "Centered vocal tract length (cm)", outer = TRUE, line=2.5)
mtext (side=2, "Centered f0 / 100 (Hz)", outer = TRUE, line=2.5)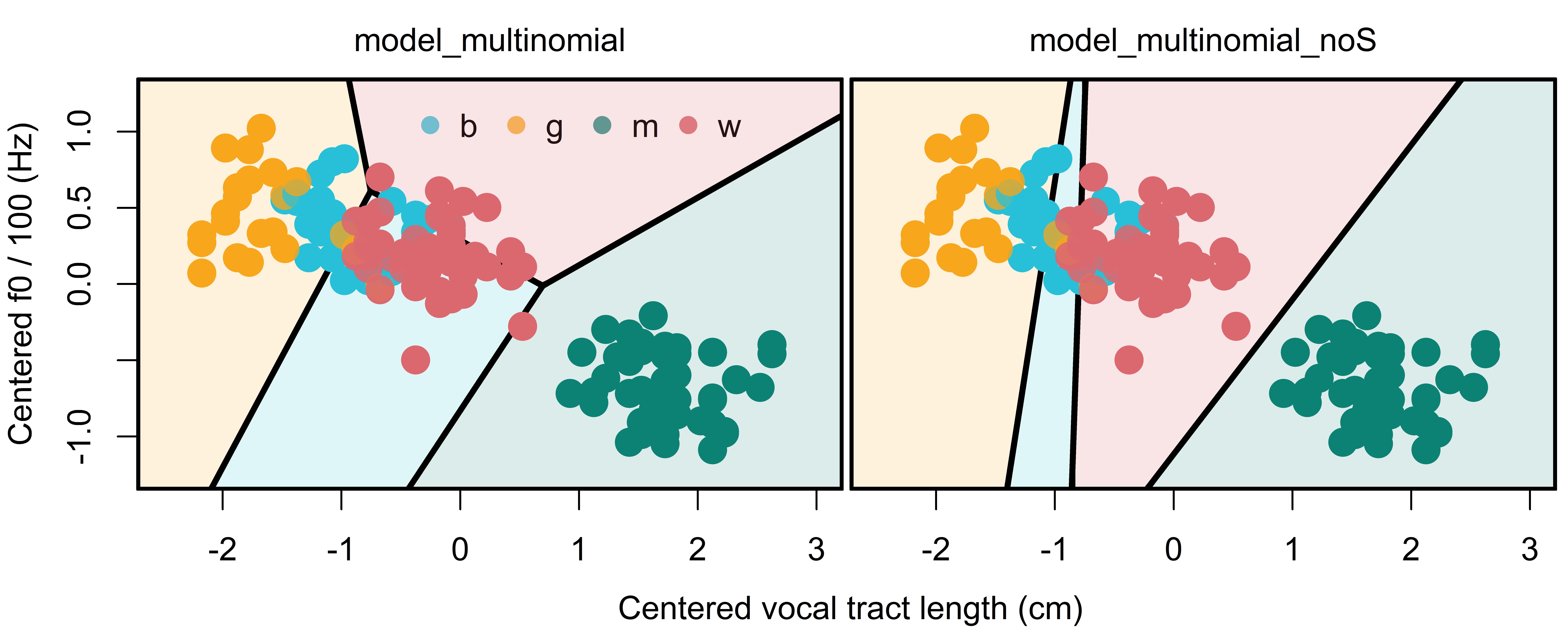
One concern with omitting the speaker effects is that our model doesn’t ‘know’ that we only had 139 speakers in our experiment. This can have the effect of artificially decreasing our credible intervals, as we saw in figure Figure 6.6. Figure 12.8 compares the fixed effects between our two models, suggesting that the omission of the speaker effects has not had a large impact on most parameters or their credible intervals. The two largest differences are for the woman category: The intercept went from slightly negative to slightly positive, and the effect for f0 went from zero to positive.
show plot code
par (mar = c(4,8,1,1))
brmplot (fixef(model_multinomial)[c(1,4,5,2,6,7,3,8,9),], las = 2,
horizontal = FALSE,col = 0, xlim = c(-5,6), xlab = "Coefficients")
abline (h = seq(1,10),lty=3,col="grey")
brmplot (fixef(model_multinomial)[c(1,4,5,2,6,7,3,8,9),], add = TRUE,
nudge = -0.2, labels = "", horizontal = FALSE, pch=16,
col = rep(bmmb::cols[3:5],each=3))
brmplot (fixef(model_multinomial_noS)[c(1,4,5,2,6,7,3,8,9),], add = TRUE,
nudge = 0.2, labels = "", horizontal = FALSE, pch=17,
col = rep(bmmb::cols[3:5],each=3))
legend (3,9.5,legend =c("With Speaker","No Speaker"),bty='n',
pch=16:17,pt.cex=1.3)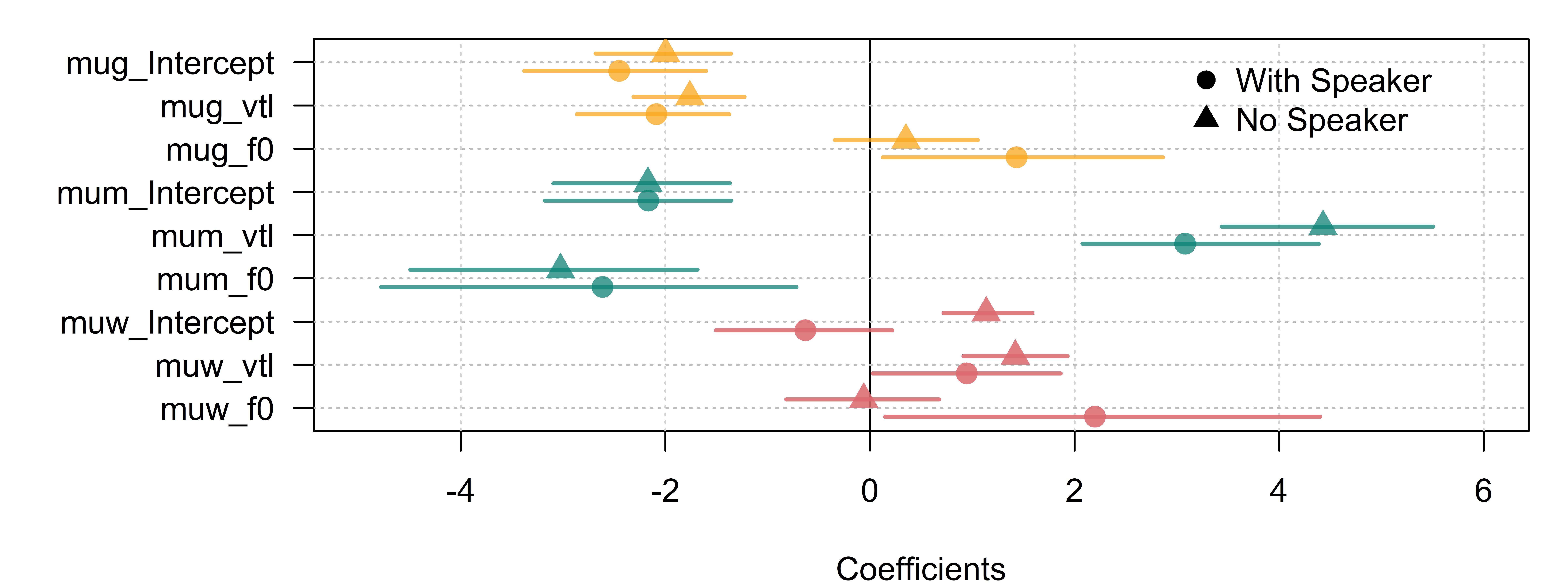
We will make new predictions for this model, with and without random effects:
multi_pred_re_noS = fitted (model_multinomial_noS)
multi_pred_noS = fitted (model_multinomial_noS, re_formula = NA)We find the maximum a posteriori speaker classification and get a label for each trial for our predictions with random effects.
predicted_noS = apply (multi_pred_re_noS[,1,],1,which.max)
predicted_category_noS = c("b","g","m","w")[predicted_noS]We calculate correct classifications overall and by category. Performance is not as good as the model with speaker effects when random effects are included (82%) but not as bad as for the same model when random effects are not included (59%). In addition, we see very good classification of all categories except for ‘boy’.
# overall correct
mean (predicted_category_noS == exp_data$C)
## [1] 0.742
# correct predictions by category
tab = xtabs (~ exp_data$C + predicted_category_noS)
diag(tab) / rowSums(tab)
## b g m w
## 0.3673 0.6737 0.9827 0.8130Importantly, we see that classification barely changes when random effects are not included, except for boy classifications which drop even below chance.
predicted_noS_no_re = apply (multi_pred_noS[,1,],1,which.max)
predicted_category_noS_no_re = c("b","g","m","w")[predicted_noS_no_re]
# overall correct
mean (predicted_category_noS_no_re == exp_data$C)
## [1] 0.7223
# correct predictions by category
tab = xtabs (~ exp_data$C + predicted_category_noS_no_re)
diag(tab) / rowSums(tab)
## b g m w
## 0.09977 0.72770 0.98268 0.9056612.2.9 Answering our research questions
We can use our models to answer the question we posed above:
(Q1) Can we use speaker f0 and VTL to predict their apparent speaker category?
Yes, we can use speaker f0 and VTL to predict apparent speaker with relatively high accuracy. If we consider a baseline correctness by chance of 25%, then our classification of 72% from two predictors (with no random effects) isn’t bad at all. However, the importance of the speaker random effects for female speakers strongly suggests that there is ‘something else’ being used to identify women as women and boys as boys, apart from VTL and f0. So, the results of our first model tells us that we should probably try to understand what that ‘something else’ is.
The speaker random effects are not particularly useful for understanding the categorization of new speakers. However, understanding what causes the variation in these random effects is. Our model tells us that even though we know there is something else to it, we can still predict categorization fairly well using just speaker VTL and f0. So, the first model may be better to really try to understand what listeners are doing when they classify individual voices, while the second model may be better to understand classification from just f0 and VTL.
12.3 Ordinal (logistic) regression
Ordinal logistic regression involves the prediction of ordered categories. Although in some senses ordinal regression is ‘simpler’ than multinomial regression, we decided to present it after multinomial regression. This is because, as discussed above, multinomial regression is mostly just a multivariate generalization of logistic regression (discussed in detail in Chapter 10). In contrast, ordinal regression, although related to logistic regression, is in many ways conceptually its own thing. We will present one way of thinking about ordinal logistic regression however, as usual, there are other ways to think of it. For more information about ordinal regression, please see Agresti (2003), Agresti (2018), and Kruschke (2014) (the same citations we provided for multinomial models, actually).
A simple example of an ordinal variable is first, second, and third place in a race. These values are categorical and not on an interval or ratio scale. For example fourth place is not ‘double’ anything with respect to second place, and the distance between first and second doesn’t necessarily represent the same difference as the difference between second and third. However, there is clearly an inherent ordering in the categories such that first < second < third < fourth < and so on. A common experimental example of ordered categorical data arises from survey data that asks listeners to respond to questions using a small number of discrete, categorical choices. For example, respondents might be asked to evaluate whether they 1) strongly agree, 2) somewhat agree, 3) are neutral, 4) somewhat disagree, or 5) strongly disagree with some proposition. This is often referred to as a Likert scale and the data resulting from this is often referred to as Likert scale data.
Likert scale data was originally meant to have response scales ranging from agreement to disagreement (as above). However, the concept of Likert scale data can be generalized to a very broad range of possible questions. For example, rather than height in centimeters, we might have asked listeners “use this 10 point scale to indicate how tall this person sounds”. This could be made into a more ‘traditional’ Likert scale by asking it in the form “this person sounds very tall” and asking people to agree or disagree on a 10 point scale. Rather than ask for binary gender judgments, we might have asked “Use this 7 point scale to judge the perceived femininity/masculinity of the speaker” (or the more traditional “this person sounds very feminine” and asking people to agree or disagree on a 7 point scale).
Although Likert scale data is sometimes treated as quantitative, there are some characteristics of this data that make an ordinal analysis a better fit. First, it is not necessarily the case that the ‘distance’ between adjacent responses is equal across all categories. We can say with absolute certainty that the difference between 150 and 151 cm is equal to the difference between 151 and 152 cm. However, is the difference between strongly agree and somewhat agree the same as the difference between somewhat agree and neutral? This is difficult if not impossible to determine. Further, the small number of outcome categories may result in subjects reserving extreme categories for extraordinary cases. What if a subject somewhat strongly agrees? There is no such option, and this subject may instead indicate they somewhat agree lest something they really strongly agree with comes along later.
Here’s what we said in Chapter 1 about when to treat a variable as quantitative:
Is the variable on a ratio or interval scale? This is a prerequisite for a quantitative value to be used as a dependent variable. An interval scale means that distances are meaningful, and a ratio scale means that 0 is meaningful.
Is the underlying value continuous? Many variables are discrete in practice due to limitations in measurement. However, if the underlying value is continuous (e.g. height, time) then this can motivate treating the measurement as a quantitative dependent variable since fractional values ‘make sense’. For example, even if you measure time only down to the nearest millisecond, a value of 0.5 milliseconds is possible and interpretable. In contrast, a value of 0.5 people is not.
Are there a large number (>50) of possible values the measured variable can take? For example a die can only take on 6 quantitative values, which is not enough.
Are most/all of the observed values far from their bounds? Human height does not really get much smaller than about 50 cm and longer than about 220 cm, so it is technically bounded. However, in most cases our observations are expected to not be clustered at the boundaries.
If survey participants were given a survey with 5 discrete numerical categories and were told that this represented some continuous value ‘agreement’, then we could perhaps argue that that 5-point Likert scale data abides by the first two considerations above. However, our data runs afoul of the bottom two considerations: There are only a small number of discrete outcomes, with nothing ‘in between’, and many of the observed values are likely to be near the boundaries and are likely to be constrained by these.
We don’t actually have any ordinal variables in our experiment, but we still wanted to provide an example of an ordinal analysis. To do this, we’re going to make a fake ordinal variable that we think is actually relatively plausible. In Figure 12.9, we see a boxplot of apparent height judgments organized by apparent speaker category, with boys and girls collapsed into one category, ‘child’. Note that apparent children are judged to be shortest, apparent women are judged to be a little taller than that, and apparent men are judged to be a little taller than women. In addition, there is not very much overlap in apparent heights between categories. Based on the above, we could potentially think of our apparent speaker categories as reflecting classifications of speakers into small (boys, girls), medium (women), and large (men). We might imagine that if we had asked listeners to identify speakers as small, medium, or large, listeners would have labelled most apparent children as small, most apparent women as medium, and most apparent men as large.
show plot code
SG = 0
SG[exp_data$C=='m'] = 3
SG[exp_data$C=='w'] = 2
SG[exp_data$C=='g'] = 1
SG[exp_data$C=='b'] = 1
x = seq (100,200,.1)
y = seq (100,200,.1)
par (mfrow = c(1,2), mar = c(.1,.1,.1,.1), oma = c(4,4,.5,.5))
boxplot (height ~ SG, data = exp_data,col=bmmb::cols[c(1,11,10)],ylim = c(100,200),
xlab = "Apparent Speaker Category",names=c("Child","Woman","Man"))
tmp = aggregate (height ~ SG, data = exp_data, FUN = mean)
abline (h = mean (tmp[1:2,2]), lwd = 2, col = bmmb::cols[15])
abline (h = mean (tmp[2:3,2]), lwd = 2, col = bmmb::cols[15])
boxplot (height ~ SG, data = exp_data,col=bmmb::cols[c(1,11,10)],
add = TRUE,xlab = "",xaxt='n')
mtext (side=1,text="Size Group", line = 2.5)
mtext (side=2,text="Apparent Height (cm)", line = 2.75)
plot (x,y, yaxt='n',type = 'l', xlim = c(120,190),ylim = c(100,200),xaxt='n')
rect (100,90,200,mean(tmp[1:2,2]), col = bmmb::cols[1],border=bmmb::cols[1])
rect (100,mean(tmp[1:2,2]),200,mean(tmp[2:3,2]), col = bmmb::cols[11],
border=bmmb::cols[11])
rect (100,mean(tmp[2:3,2]),200,400, col = bmmb::cols[10],border=bmmb::cols[10])
lines (x,y, lwd=4, col = bmmb::cols[2])
points (c(140,160,180),c(140,160,180),pch=16,col=bmmb::cols[2],cex=2)
abline (h = mean (tmp[1:2,2]), lwd = 6, col = bmmb::cols[15])
abline (h = mean (tmp[2:3,2]), lwd = 6, col = bmmb::cols[15])
text (118,c(140,163,195), c("Small", "Medium", "Large"),cex=1.1,pos=4)
axis (side=1, at = seq(110,190,length.out=6), labels = 11:16)
mtext (side=1,text="Vocal tract length (cm)", line = 2.75)
The right plot in Figure 12.9 presents the y axis divided into three sections. The boundaries between sections are halfway between the means of apparent children and adult females (155.7 cm), and apparent adult females and adult males (170 cm). We can think of these sections as reflecting the expected categorization of speakers into small, medium, or large based on their apparent height. We can see how this might work using the code below. First, we convert our observed speaker classifications into a new variable that we will treat as ordinal. This variable is size group (SG), and it has the values 1 (small), 2 (medium), and 3 (large).
SG = 0
SG[exp_data$C=='b' | exp_data$C=='g'] = 1
SG[exp_data$C=='w'] = 2
SG[exp_data$C=='m'] = 3Then, we create a new variable that will hold our size group predictions, SG_hat. After this, we set this variable to 1 if apparent height is less than the boundary between small and medium (155.7 cm), to 2 if apparent height is greater than the boundary between small and medium (155.7), but less than the boundary between medium and large (170 cm), and to 3 if it is greater than the boundary between medium and large (170 cm).
SG_hat = exp_data$height * 0
SG_hat[exp_data$height <= 155.7] = 1
SG_hat[exp_data$height > 155.7 & exp_data$height <= 170] = 2
SG_hat[exp_data$height >= 170] = 3This relatively primitive ‘model’ is able to correctly predict size group responses in 75% of cases.
mean (SG == SG_hat)
## [1] 0.7549The line in the right plot in Figure 12.9 reflects the relationship between speaker VTL and apparent height. We know from earlier chapters that a longer speaker VTL is associated with taller apparent speakers. Let’s assume for the sake of simplicity that this relationship is causal and that there is a direct connection between speaker VTL and apparent height. This means that when we hear a voice, we can be thought of as sliding along the line in the plot based on the VTL of the speaker. Then, based on the apparent height value at a given x axis location, we can arrive at a size group classification based on the method above. Note that in this scenario we were not collecting or observing apparent speaker height but rather predicting size group using VTL. However, we assume that apparent height underlies this process and connects the two variables we do observe. By assuming that apparent height was driving this process but not trying to measure it directly we are treating it as a latent variable.
The latent variable was presented as apparent height, however, the latent variable could have had any name and used any other units. For example, in our situation we can think of this variable as ‘size’ or ‘bigness’, but its name is not important, and neither is us having a rock-solid conceptual grasp of what it is exactly. Instead, all we need to understand is that we think there is some latent variable, something like ‘bigness’, that allows listeners to classify speakers into small, medium, and large groups based on their voices.
The right side of Figure 12.10 is labelled “bigness” to correspond to our hypothetical latent variable. This variable is just apparent height divided by 50, but the point is that the values of this axis are entirely arbitrary. Notice that the change in axis units has absolutely no effect on the way our conceptual model works. If the latent variable units are 50 times smaller than some other variable, that just means the slope of the line relating x to y is 50 times smaller and the boundaries between categories are 50 times closer. The same logic applies to the relationship between our latent dependent variable and our predictors.
show plot code
tmp = aggregate (height ~ SG, data = exp_data, FUN = mean)
x = seq (-35,35,.1)
y = dlogis(x,0,5)
x_2 = seq (100,230,.1)
y_2 = x_2
par (mfrow = c(1,1), mar = c(4,4,.1,4), oma = c(.5,.5,.51,.51))
#layout (mat = t(c(1,2)), widths = c(.6,.4))
plot (x,y, yaxt='s',type = 'l', xlim = c(120,190),ylim = c(105,215),
xlab = "Vocal-tract length (cm)",xaxt='n',ylab="Apparent Height (cm)")
rect (100,90,200,mean(tmp[1:2,2]), col = bmmb::cols[1],border=bmmb::cols[1])
rect (100,mean(tmp[1:2,2]),200,mean(tmp[2:3,2]), col = bmmb::cols[11],border=bmmb::cols[11])
rect (100,mean(tmp[2:3,2]),200,400, col = bmmb::cols[10],border=bmmb::cols[10])
lines (x_2,y_2, lwd=4, col = bmmb::cols[2])
axis (side=4, at = seq(120,200,20), labels = seq(120,200,20)/5)
spot = 160
lines (-y*150+spot, x+spot, ylim = c(120,190), xlim = c(-.35,0), xaxs='i',type='l',lwd=2)
lines (rep(spot,length(x)),x+spot,type="l",lwd=2)
points (spot,spot,pch=16,col=bmmb::cols[2],cex=2)
spot = 140
lines (-y*150+spot, x+spot, ylim = c(120,190), xlim = c(-.35,0), xaxs='i',type='l',lwd=2)
lines (rep(spot,length(x)),x+spot,type="l",lwd=2)
points (spot,spot,pch=16,col=bmmb::cols[2],cex=2)
spot = 180
lines (-y*150+spot, x+spot, ylim = c(120,190), xlim = c(-.35,0), xaxs='i',type='l',lwd=2)
lines (rep(spot,length(x)),x+spot,type="l",lwd=2)
points (spot,spot,pch=16,col=bmmb::cols[2],cex=2)
text (118,c(140,163,195), c("Small", "Medium", "Large"),cex=1.1, pos=4)
abline (h = mean (tmp[1:2,2]), lwd = 5, col = bmmb::cols[15])
abline (h = mean (tmp[2:3,2]), lwd = 5, col = bmmb::cols[15])
axis (side=1, at = seq(110,190,length.out=6), labels = 11:16)
lines (x,y, lwd=4, col = bmmb::cols[2])
mtext (side=4, "Bigness", line=2.2)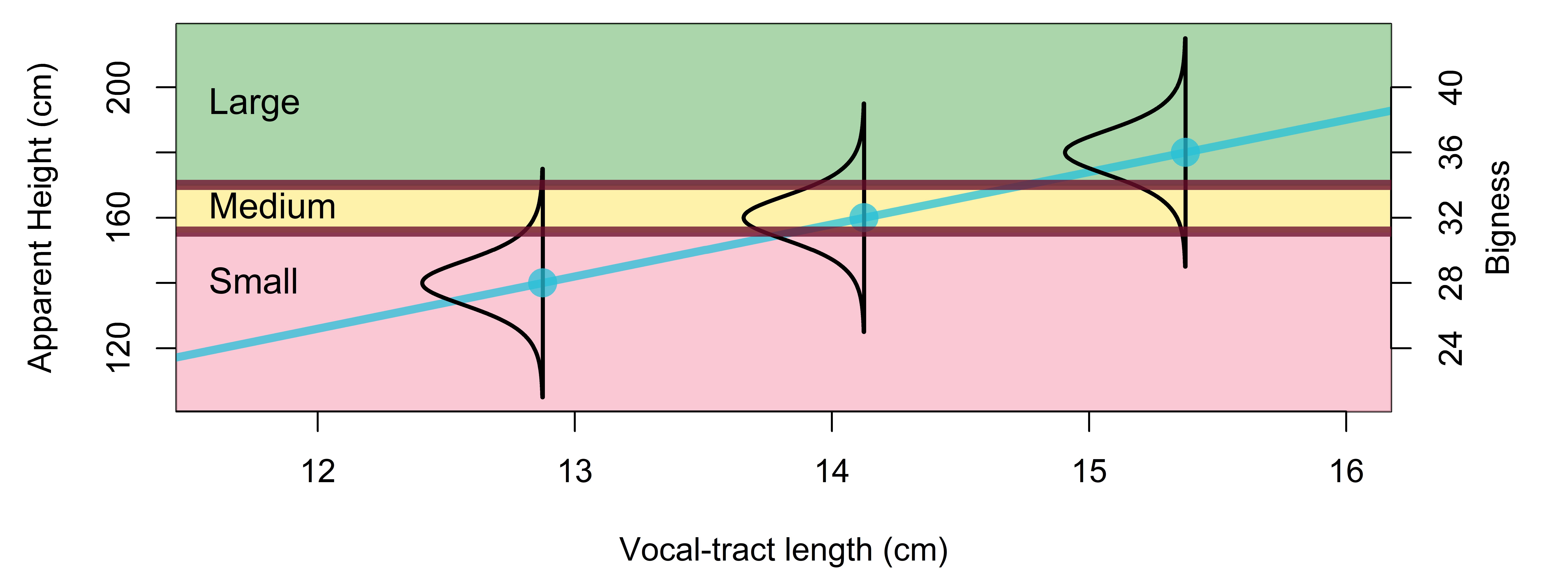
Our ‘model’ so far would do a reasonable job of predicting classification but contains no random component. As a result, given a certain speaker VTL it would always predict the same size group response. In Figure 9.1, we showed that bivariate linear regression with normally-distributed errors can be thought of as a normal distribution sliding along a line based on the value of the dependent variable. The y axis value of the line at an x axis location tells you the expected value of y for that value of x. However, we will hardly (if ever) observe the expected value exactly. Instead, we assume that we have normal errors centered around our expected values.
We show a similar situation in Figure 12.10, with a probability distribution sliding along a line. However, in the case of ordinal regression the values that this distribution generates are not the observed values of the ordinal dependent variable, but rather random values of the latent variable (e.g. ‘bigness’). The observed ordinal variable is then based on the value of the latent variable with respect to the boundaries in the space (the horizontal lines in the figure). In this chapter, we will use a logistic distribution as the error distribution (more detail on this in the next section), hence the name, ordinal logistic regression. We will do this because it is commonly used and has some useful characteristics, but a basically equivalent model can be constructed using a normal distribution to represent randomness in the latent variable.
Ordinal regression estimates the location of boundaries between categories along the latent variable. It also tries to predict the expected value of the latent variable for each observation, based on the combination of the dependent variables. For example, in Figure 12.10 the boundaries are at 31 and 34 bigness, and you have an expected bigness of around 32 for a vocal-tract length of 14.1. We can think of it this way: Given a VTL of 14.1 we know how big people will sound in general (32), but these things are fuzzy and necessarily noisy. So actually, there is a distribution of expected perceived/apparent ‘bigness’ given our predictors. Given the actual value of the latent variable for a trial (including the random component), the ordinal response is based on the value of the latent variable with respect to the estimated category boundaries. So, for a fixed expected value of 32, sometimes the distribution might ‘generate’ a bigness value of 28, resulting in a response of small. For another observation with the same expected value, the value of the latent variable may be 36, meaning the speaker will be identified as large.
12.3.1 Cumulative distribution functions
In order to understand ordinal regression, we need to talk about cumulative distribution functions first. We haven’t explicitly talked about cumulative distribution functions yet, though we did see one in Chapter 10 when it was referred to as the inverse logit function (more on this in a moment). To this point we’ve been focused on what are called probability density functions (PDF), functions that assign a density to different values of a variable. Examples of PDFs are given in the top row of Figure 12.11. Cumulative distribution functions (CDF) tell you the probability that a variable will be less than or equal to some value. Since a PDF has an area under the curve equal to 1 (representing the total probability of the variable), a CDF tells you how much of that area (out of 1) is to the left of the value, and has a range from 0 to 1.
For example, in the left column of Figure 12.11 we see a standard normal distribution with a mean of 0 and a standard deviation of one. We know that the normal distribution is symmetrical about its mean so that half the area under the curve of this density must be below 0. If we look at the bottom row of the first column, we can see the CDF corresponding to the PDF in the top row. If we look at the value of this function at x=0, we see that it is exactly equal to 0.5. In other words, the cumulative function tells us that the standard normal has exactly half its mass at values less than x=0, i.e. the probability of observing a value of x that is less than or equal to 0, the mean, is exactly 0.5.
show plot code
par (mfcol = c(2,3), mar = c(.1,.2,.11,.2), oma = c(2.5,4,3,.5))
x = seq(-5,5,.01)
plot (x, dnorm (x), type = 'l', xaxt='n', xaxs='i',lwd=4,col=bmmb::cols[5],yaxt='n')
mtext (side=2,"Density", line=2.5)
mtext (side=3,"Standard Normal", line=1)
abline (v = 0,lty=3,col="grey",lwd=3)
plot (x, pnorm (x), type = 'l', xaxs='i',lwd=4,col=bmmb::cols[5])
mtext (side=2,"Cumulative Probability", line=2.5)
segments (-6,.5,0,.5,lty=3,col="grey",lwd=3)
segments (0,-5,0,.5,lty=3,col="grey",lwd=3)
text (0.1,.2,"x=0", cex = 1.5,pos=4)
text (-3,.6,"y=0.5", cex = 1.5,pos=4)
x = seq(-10,10,.01)
plot (x, dlogis (x), type = 'l', xaxt='n', yaxt='n', xaxs='i',lwd=5,col=bmmb::cols[2])
mtext (side=3,"Logistic", line=1)
abline (v = 2,lty=3,col="grey",lwd=3)
plot (x, plogis (x), type = 'l', yaxt='n', xaxs='i',lwd=5,col=bmmb::cols[2])
segments (-16,.88,2,.88,lty=3,col="grey",lwd=3)
segments (2,-5,2,.88,lty=3,col="grey",lwd=3)
text (2.1,.2,"x=2", cex = 1.5,pos=4)
text (-7,.8,"y=0.88", cex = 1.5,pos=4)
x = seq(-10,10,.01)
plot (x, dlogis (x), type = 'l', xaxt='n', yaxt='n', xaxs='i',lwd=5,col=bmmb::cols[1])
mtext (side=3,"Logistic", line=1)
abline (v = c(2,-1),lty=3,col="grey",lwd=3)
plot (x, plogis (x), type = 'l', yaxt='n', xaxs='i',lwd=5,col=bmmb::cols[1])
segments (-16,.88,2,.88,lty=3,col="grey",lwd=3)
segments (-16,.27,-1,.27,lty=3,col="grey",lwd=3)
segments (2,-5,2,.88,lty=3,col="grey",lwd=3)
segments (-1,-5,-1,.27,lty=3,col="grey",lwd=3)
text (2.1,.2,"x=2", cex = 1.5,pos=4)
text (-7,.8,"y=0.88", cex = 1.5,pos=4)
text (-5.11,.1,"x= -1", cex = 1.5,pos=4)
text (-7,.35,"y=0.27", cex = 1.5,pos=4)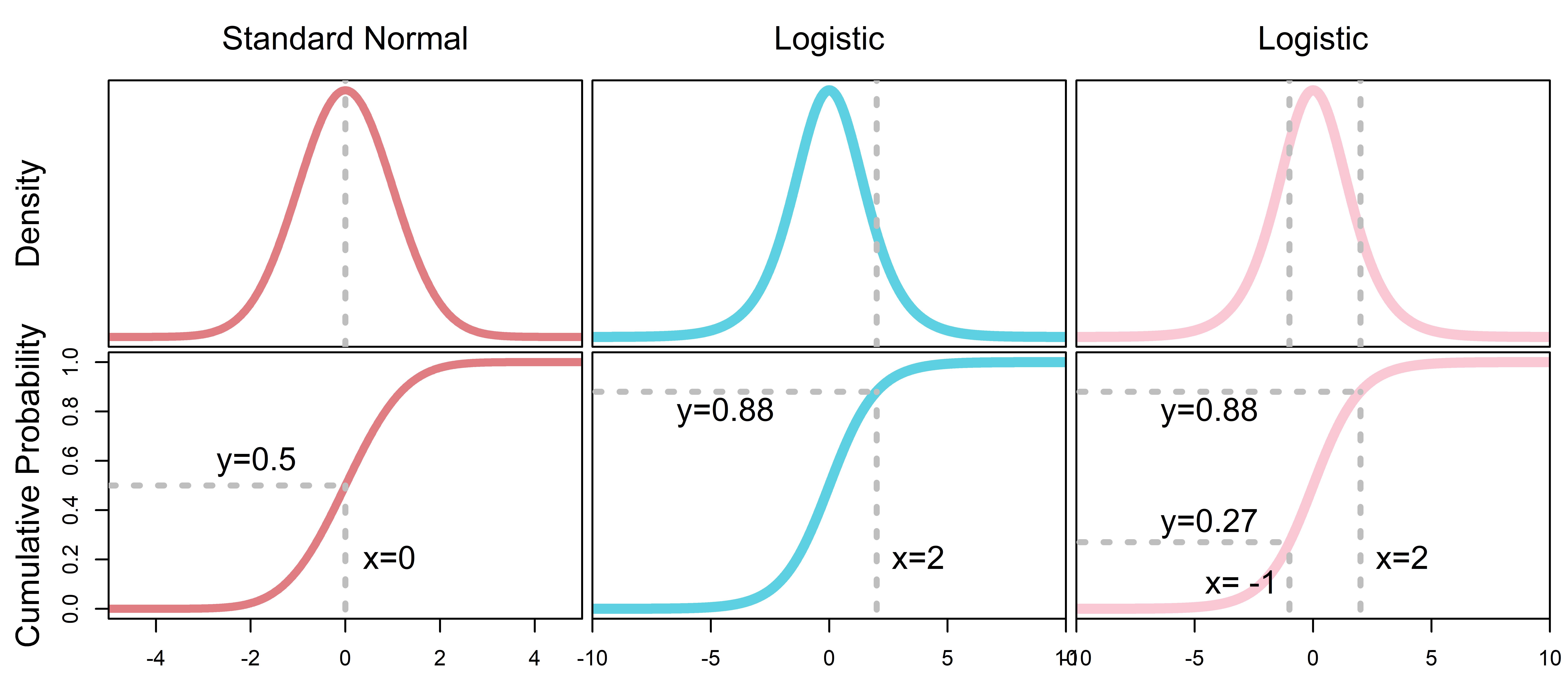
We can use the pnorm function (discussed in Chapter 2) to get the value of the normal CDF for any value of x. Below we calculate this for x=0, x= -2, and x=2. See that in each case, the output of the function corresponds to the y axis value of the CDF at that x axis location (bottom left, Figure 12.11).
# cumulative distribution at x = -2, 0, 2
pnorm (c(-2,0,2),0,1)
## [1] 0.02275 0.50000 0.97725The CDF is the integral of the PDF, and the PDF is the derivative of the CDF. We’ve already explained the integral part of this: As you move left to right, the value of the CDF equals the cumulative (i.e. added up total) area under the curve (i.e. probability) in the PDF to that point. To say that the PDF is the derivative of the CDF means that the value of the PDF reflects the rate of change (i.e. the slope) in the CDF for different values of x. Imagine you were in a tiny car driving along the standard normal CDF in Figure 12.11, left to right. As you drive along around -4 the ‘terrain’ is flat and the value of the density is near 0. As you near -2 the slope of the CDF begins to increase, as does the value of the PDF. The value of the PDF is largest at x=0, telling us that the slope of the CDF is greatest at that point. As we drive past x=0 in the CDF, we see a gradual decrease in the slope until we more or less reach ‘flat ground’ again at just past x=2. This is reflected by the gradual decrease in the value of the PDF between x=0 and x=2.
In the middle column of Figure 12.11 we see a logistic distribution. The logistic distribution is a two-parameter distribution similar in shape to the normal distribution but with ‘fatter/heavier’ tails (like the t-distribution). The logistic distribution has a location parameter (\(\mu\), equal to the mean) that determines its location along the number line, and a scale parameter (\(s\), equal to the standard deviation times \(\sqrt{3}/\pi\)) that is positively related to the ‘width’ of the density function. In the bottom row of the middle column, we see the CDF of the logistic density. It turns out that the inverse logit function we use as the link function for logistic regression is simply the CDF of a logistic distribution with a mean of 0 and a scale of 1 (i.e. a standard logistic distribution). This is also why you might commonly see the inverse logit function referred to as the logistic function. We avoided using this name for the function to avoid confusion.
In the middle column of Figure 12.11 we see that we can use the same approach we used for our normal distribution to calculate values of the CDF of our logistic distribution. Below, we use the plogis function to calculate the probability of observing a value greater than or equal to 2 form that distribution.
plogis (2,0,1)
## [1] 0.8808In the right column of Figure 12.11 we’ve placed two vertical lines at x= -1 and x=2. We use the code below to calculate the area under the curve to the left of each line.
# area left of first line
plogis (-1,0,1)
## [1] 0.2689
# area left of second line
plogis (2,0,1)
## [1] 0.8808At this point, the connection between CDFs and ordinal regression may be clear: We can use a set of boundaries and the CDF of a probability distribution to calculate the probability of observing different values of the latent variable, within certain intervals. For example, in the bottom right plot of Figure 12.11, if 0.27 falls to the left of the first line and 0.88 falls to the left of the second line, then 0.61 must fall between the first and second lines. Thus, we have effectively divided the distribution into three parts and can discuss the probability that the observation of the variable will fall within each part.
# area between first and second line
plogis (2,0,1)-plogis (-1,0,1)
## [1] 0.6119In Figure 12.12, the boundaries and distributions from Figure 12.10 have been rotated clockwise 90 degrees so that our latent variable (the arbitrarily named ‘bigness’) varies along the x axis. The distributions presented in the figure have means of 28, 32, 36, and scales of 1. The probabilities in each subsection of each plot represent the predicted probability of observing size group responses of 1, 2, and 3 for each distribution. These probabilities were calculated based on the area under the curve of the distributions falling between different intervals of the latent variable space.
show plot code
x = seq (20,44,.01)
y = dlogis(x,30,1)
x_2 = seq (-4,4.4,.05)
y_2 = x_2
par (mfrow = c(3,1), mar = c(.5,1,.5,1), oma = c(4,.5,.51,.51))
plot (x+3.2,y*2, yaxt='s',type = 'n', xlim = c(22,42),ylim = c(0.02,.8),
xlab = "",xaxt='n',ylab="",yaxt = 'n')
rect (1,0,31,1, col = bmmb::cols[1],border=bmmb::cols[1])
rect (31,0,34,1, col = bmmb::cols[11],border=bmmb::cols[11])
rect (34,0,50,1, col = bmmb::cols[10],border=bmmb::cols[10])
lines (x-2,y*2.5, xaxs='i',type='l',lwd=2)
abline (v = c(31,34),lwd=5,col=bmmb::cols[15])
abline (v = 28, lty=3)
text (25, .45, "P(SG=1)=0.95", cex=1.1)
text (32.5, .3, "P(SG=2)=0.04", cex=1.1)
text (40, .3, "P(SG=3)=0.01", cex=1.1)
plot (x+3.2,y*2, yaxt='s',type = 'n', xlim = c(22,42),ylim = c(0.02,.8),
xlab = "Bigness",xaxt='n',ylab="",yaxt = 'n')
rect (1,0,31,1, col = bmmb::cols[1],border=bmmb::cols[1])
rect (31,0,34,1, col = bmmb::cols[11],border=bmmb::cols[11])
rect (34,0,50,1, col = bmmb::cols[10],border=bmmb::cols[10])
lines (x+2,y*2.5, type = 'l', lwd=2)
abline (v = c(31,34),lwd=5,col=bmmb::cols[15])
abline (v = 32, lty=3)
text (25, .3, "P(SG=1)=0.26", cex=1.1)
text (32.5, .2, "P(SG=2)=0.61", cex=1.1)
text (40, .3, "P(SG=3)=0.12", cex=1.1)
plot (x+3.2,y*2, yaxt='s',type = 'n', xlim = c(22,42),ylim = c(0.02,.8),
xlab = "Bigness",xaxt='n',ylab="",yaxt = 'n')
rect (1,0,31,1, col = bmmb::cols[1],border=bmmb::cols[1])
rect (31,0,34,1, col = bmmb::cols[11],border=bmmb::cols[11])
rect (34,0,50,1, col = bmmb::cols[10],border=bmmb::cols[10])
axis (side=1, at = seq(24,40,4),cex.axis=1.2)
lines (x+6,y*2.5, type = 'l', lwd=2)
abline (v = c(31,34),lwd=5,col=bmmb::cols[15])
abline (v = 36, lty=3)
text (25, .3, "P(SG=1)=0.01", cex=1.1)
text (32.5, .35, "P(SG=2)=0.11", cex=1.1)
text (40, .3, "P(SG=3)=0.88", cex=1.1)
mtext (side=1,"Bigness", line=2.75)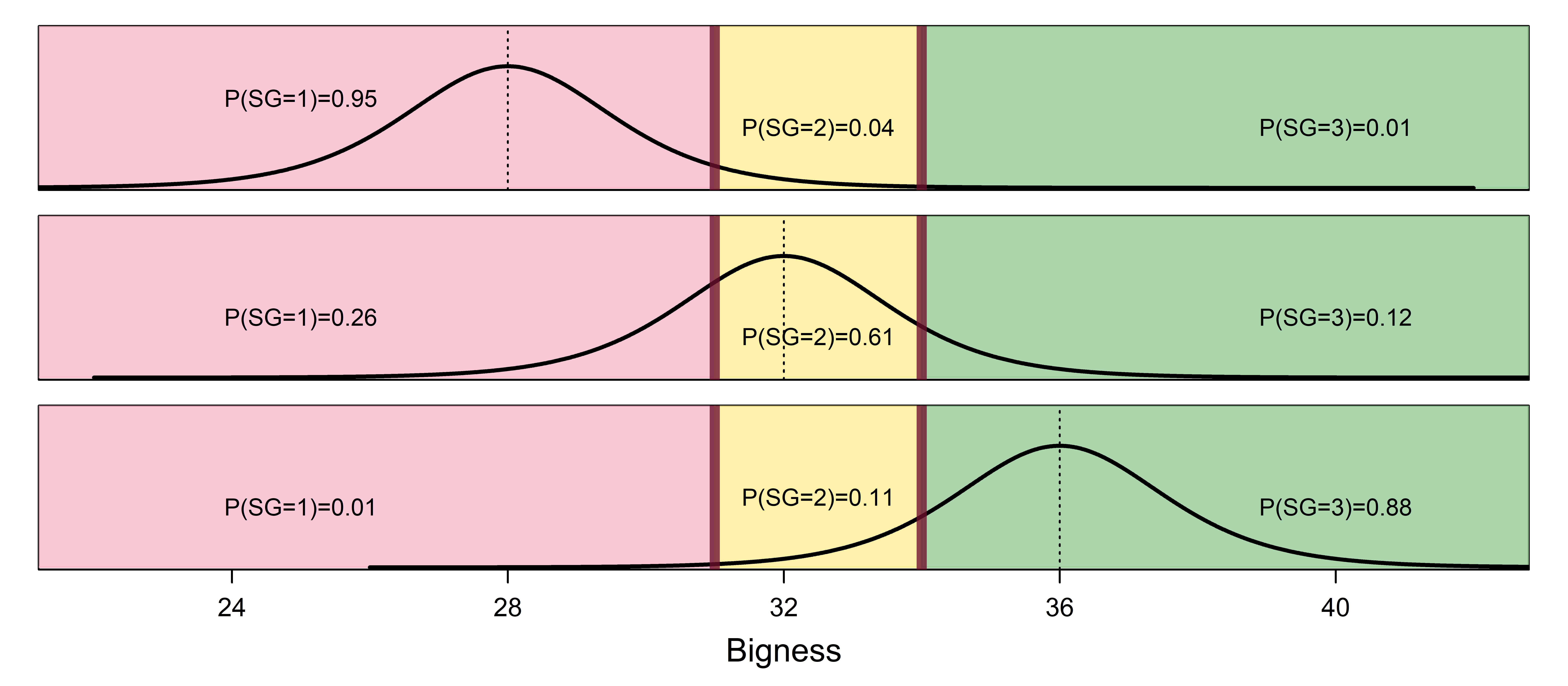
For example, below we calculate the probability of observing each size group based on the distribution in the middle plot of Figure 12.12, a latent variable distribution with a mean of 32 and a scale of 1. First, we find the probability of observing a bigness value less than 31 (the first boundary). This tells us the probability of observing a response of size group 1 for this expected value of bigness (P(SG=1)=0.26). Then, we again find the probability of a bigness value less than 31 (the first boundary), and subtract this from the probability of a bigness value less than 34 (the second boundary). This operation gives us the probability of observing a bigness value between the first and second boundaries, i.e. the probability of observing a medium size group response (P(SG=2) = 0.88 - 0.26). Finally, to find the probability of a large size group response we find the probability of a bigness value less than 34 and subtract this from 1. Since there are no more boundaries above the largest response category, the entire probability above the highest threshold must correspond to the final category.
# probability of observing category 1
plogis (31,32,1)
## [1] 0.2689
# probability of observing category 2
plogis (34,32,1) - plogis (31,32,1)
## [1] 0.6119
# probability of observing category 3
1 - plogis (34,32,1)
## [1] 0.1192This conceptualization of our model leads to both hard classification and soft classification. An ordinal regression model will predict, for every observation, an expected value of the dependent variable based on the values of the predictors. This value can then be used to find response probabilities for each category. A hard classification classifies each observation into a predicted category, 1, 2, or 3, based on the value of the highest predicted probability across the categories. A soft classification provides the probability that an observation is classified into one or more categories. For example, in the middle plot of Figure 12.12, it may be useful to say that the most probable classification is of a ‘medium’ speaker. However, it may be just as, or more, useful to indicate that the probability of this response was only 0.61, making the observation of a small response (P=0.26) not unlikely.
12.3.2 Data and research questions
We load our packages and data below. We also add a new variable, SG (size group), which will act as our dependent variable. This variable represents children with a 1, adult women with a 2, and adult males with a 3. We don’t need to do anything special to make this variable ‘ordinal’, but we are using three consecutive integers to represent the categories to make things simpler. After this, we process our quantitative predictors in the same way as in the previous chapters.
library (brms)
library (bmmb)
data (exp_data)
# new dependent variable: Size Group
SG = 0
SG[exp_data$C=='g'] = 1
SG[exp_data$C=='b'] = 1
SG[exp_data$C=='w'] = 2
SG[exp_data$C=='m'] = 3
exp_data$SG = SG
# preparation of quantitative predictors as in previous chapters
exp_data$vtl_original = exp_data$vtl
exp_data$vtl = exp_data$vtl - mean (exp_data$vtl)
exp_data$f0_original = exp_data$f0
exp_data$f0 = exp_data$f0 - mean(exp_data$f0)
exp_data$f0 = exp_data$f0 / 100We’re going to keep our research questions relatively simple this time, as this is mostly a demonstration of an ordinal regression analysis and not a ‘real’ ordinal variable:
(Q1) Can we predict size group responses for a speaker given their f0 and VTL?
12.3.3 Description of the model
To fit a model predicting apparent size group from speaker f0 and VTL, we can use the formula below:
SG ~ vtl+f0 + (vtl+f0|L) + (1|S)
This model formula is very much like ones we’ve used several times in earlier chapters. The formula says “predict size group using a plane based on voice f0 and VTL, allow for listener-specific planes, and speaker-specific adjustments to the height of the plane”. However, the relationship it represents between our predictors and our dependent variables is substantially different to what we’ve seen before. Let’s begin with the relationship between the independent variables and the expected value of the latent variable (i.e. the mean of our logistic distribution), presented in equation Equation 12.16:
\[ \begin{equation} \begin{split} \mu_{[i]} = a_{[i]} + b_{[i]} \cdot \mathrm{vtl}_{[i]} + c_{[i]} \cdot \mathrm{f0}_{[i]} \end{split} \end{equation} \tag{12.16}\]
We model this latent variable as being distributed according to a logistic distribution with a mean of \(\mu\) and a scale parameter equal to 1, as in equation Equation 12.17. Actually, for reasons that we won’t get into, for ordinal regression brm uses a parameter called the discrimination parameter, which is the inverse of the scale. However, since we are using a scale of 1 we can ignore this for now (since 1/1=1).
\[ \begin{equation} \begin{split} z_{[i]} \sim \mathrm{Logistic} (\mu_{[i]}, 1) \end{split} \end{equation} \tag{12.17}\]
Our observed (dependent) ordinal category is then predicted based on the value of the latent variable (\(z_{[i]}\)) for that trial, relative to the model boundaries, called thresholds, or cutoffs (\(\theta_{[1]},\theta_{[2]}\)). We see this in Equation 12.18 below.
\[ \begin{equation} \begin{split} SG_{[i]} = \begin{cases} 1 \; \mathrm{if} \; z_{[i]} \leq \theta_1 \\ 2 \; \mathrm{if} \; \theta_{[1]} < z_{[i]} \leq \theta_{[2]} \\ 3 \; \mathrm{if} \; \theta_{[2]} \leq z_{[i]} \\ \end{cases} \end{split} \end{equation} \tag{12.18}\]
The above says: “SG is equal to 1 if the latent variable is below the first threshold, 2 if it’s between the first and second thresholds, and 3 if it is above the second threshold. Here is a full description of our model:
\[ \begin{equation} \begin{split} SG_{[i]} = \begin{cases} 1 \; \mathrm{if} \; z_{[i]} \leq \theta_{[1]} \\ 2 \; \mathrm{if} \; \theta_{[1]} < z_{[i]} \leq \theta_{[2]} \\ 3 \; \mathrm{if} \; \theta_{[2]} \leq z_{[i]} \\ \end{cases} \\\\ z_{[i]} \sim \mathrm{Logistic} (\mu_{[i]}, 1)\\ \\ \mu_{[i]} = a_{[i]} + b_{[i]} \cdot \mathrm{vtl}_{[i]} + c_{[i]} \cdot \mathrm{f0}_{[i]} \\ a_{[i]} = L_{[\mathsf{L}_{[i]}]} \\ b_{[i]} = VTL + VTL \colon L_{[\mathsf{L}_{[i]}]} \\ c_{[i]} = F0 + F0 \colon L_{[\mathsf{L}_{[i]}]} \\ \\ \textrm{Priors:} \\ S_{[\bullet]} \sim \mathrm{Normal}(0,\sigma_{S}) \\ \begin{bmatrix} L_{[\bullet]} \\ VTL \colon L_{[\bullet]} \\ F0 \colon L_{[\bullet]} \end{bmatrix} \sim \mathrm{MVNormal} \left(\, \begin{bmatrix} 0\\ 0 \\ 0 \\ \end{bmatrix}, \mathrm{\Sigma} \right) \\ \\ \theta_{[1]}, \theta_{[2]} \sim \mathrm{t}(3, 0, 3) \\ VTL, F0 \sim \mathrm{t}(3, 0, 3) \\ \sigma_{L}, \sigma_{VTL \colon L}, \sigma_{F0 \colon L}, \sigma_{S} \sim \mathrm{t}(3, 0, 3) \\ R \sim \mathrm{LKJCorr} (2) \end{split} \end{equation} \tag{12.19}\]
And here is a plain English, verbal description of our model:
We are modelling an ordinal variable, size group (\(SG\)), with possible outcomes of small (1), medium (2), and large (3). The value expected for any given trial is based on the value of a latent variable (\(z\)) in relation to two thresholds (\(\theta_{[1]},\theta_{[2]}\)), which were estimated from the data. Our latent variable is modeled as coming from a logistic distribution with a scale of 1 and a mean that varies from trial to trial. The mean of these distributions varies based on the combination of ‘main’ (e.g. \(VTL\)) and listener-dependent (e.g. \(VTL \colon L\)) effects for vocal tract length and f0, and a listener-dependent intercept. The speaker intercept (\(S\)) terms were drawn from a normal distribution with a mean of zero and a standard deviation estimated from the data. The listener random effects were drawn from a multivariate normal distribution with means of zero and a correlation matrix (R) estimated from the data. All other effects were treated as ‘fixed’ and drawn from prior distributions appropriate for their expected range of values.
Notice that the model in Equation 12.19 does not contain a term representing the overall model intercept. This is because the important thing for our model is the distance between the expected value of the latent variable and the thresholds, and not the value of the latent variable itself. For example, imagine an ‘intercept-only’ model (Chapter 4) that had intercept = 3 and thresholds of 1 and 5. The behavior of this model would be exactly analogous to a model with an intercept of 13 and thresholds of 11 and 15. In fact, there are an infinite number of models that maintain this structure, one for each possible value of the intercept. Under these conditions finding the ‘best’ model is an impossible task (i.e. the model is not identifiable, see Section 8.8). To resolve this issue, the overall intercept in these models is usually set to zero and not modeled, and instead only the \(J-1\) thresholds are modeled (for \(J\) categories).
Although ordinal models do not usually estimate the overall intercept, they can estimate listener-dependent (and speaker-dependent) intercepts, and our model in equation does contain these. Since the boundaries are shared by all groups, using group-dependent intercepts can be thought of as sliding the thresholds up and down the number line by different amounts for each level of a grouping factor (e.g. listener), or as moving the predicted values by the same amount. This sort of adjustment can have a meaningful effect on predictions, unlike changes in the overall model intercept outlined above.
12.3.4 Fitting and interpreting the model
To fit an ordinal logistic model, you set family=cumulative when fitting your model. The logistic distribution is the default error distribution for these sorts of models, but other distributions such as the normal distribution can be used.
# Fit the model yourself
model_ordinal =
brms::brm (SG ~ vtl+f0 + (vtl+f0|L) + (1|S), data=exp_data,
family="cumulative", chains=4, cores=4, warmup=1000,
iter = 5000, thin = 4,
prior = c(set_prior("student_t(3, 0, 3)", class = "Intercept"),
set_prior("student_t(3, 0, 3)", class = "b"),
set_prior("student_t(3, 0, 3)", class = "sd"),
set_prior("lkj_corr_cholesky (2)", class = "cor")))# Or download it from the GitHub page:
model_ordinal = bmmb::get_model ("12_model_ordinal.RDS")We can inspect the short summary to see the information it contains:
bmmb::short_summary(model_ordinal)
## Formula: SG ~ vtl + f0 + (vtl + f0 | L) + (1 | S)
##
## Group-Level Effects:
## ~L (Number of levels: 15)
## Estimate Est.Error l-95% CI u-95% CI
## sd(Intercept) 0.54 0.15 0.30 0.89
## sd(vtl) 0.45 0.20 0.07 0.89
## sd(f0) 0.67 0.40 0.04 1.55
## cor(Intercept,vtl) 0.07 0.33 -0.58 0.67
## cor(Intercept,f0) 0.14 0.36 -0.58 0.79
## cor(vtl,f0) -0.11 0.37 -0.78 0.62
##
## ~S (Number of levels: 139)
## Estimate Est.Error l-95% CI u-95% CI
## sd(Intercept) 1.27 0.14 1.02 1.57
##
## Population-Level Effects:
## Estimate Est.Error l-95% CI u-95% CI
## Intercept[1] -1.86 0.22 -2.32 -1.44
## Intercept[2] 2.58 0.24 2.13 3.07
## vtl 3.16 0.29 2.63 3.76
## f0 -1.78 0.60 -2.98 -0.58
##
## Family Specific Parameters:
## Estimate Est.Error l-95% CI u-95% CI
## disc 1 0 1 1In the population-level effects, we see the two estimated thresholds Intercept[1] and Intercept[2] representing \(\theta_{[1]}\) and \(\theta_{[2]}\) above. We also see our ‘main’ effects for VTL and f0 in this section. Our results indicate that VTL is positively related to a larger size group response, while f0 is negatively related to this (no surprises there). In the group-level effects section, note that we get listener-dependent intercepts, VTL, and f0 effects. The intercepts correspond to the \(L\) terms in the model described in Equation 12.19, and do not refer to listener-dependent thresholds/cutoffs. Below we make ‘fixed effect’ predictions using our model. These predictions omit the random effects structure to let us see how well our fixed effects can predict our data. In addition, we set scale="linear" so that we get predictions of the latent variable rather than the probability of category membership.
# make latent variable predictions
predictions_latent = fitted (model_ordinal,scale="linear", re_formula = NA)
# inspect first 6 predictions
head (predictions_latent)
## Estimate Est.Error Q2.5 Q97.5
## [1,] -5.000 0.3558 -5.742 -4.330
## [2,] -4.545 0.3958 -5.330 -3.783
## [3,] -5.864 0.4669 -6.817 -4.994
## [4,] -5.645 0.3486 -6.376 -4.978
## [5,] -4.728 0.2945 -5.340 -4.171
## [6,] -7.115 0.5044 -8.168 -6.163Below, we predict our latent variable using our posterior mean fixed effects. We do this just to show that it does, in fact, result in the expected predictions. Below, we get the posterior mean model fixed effects and use these to come up with an expected value for our latent variable for each observation (mu).
# get fixed effects
fixed_effects = brms::fixef(model_ordinal)
threshold_1 = fixed_effects[1,1]
threshold_2 = fixed_effects[2,1]
vtl_slope = fixed_effects[3,1]
f0_slope = fixed_effects[4,1]
# get expected values
mu = exp_data$vtl * vtl_slope + exp_data$f0 * f0_slopeWe can see that the prediction above results in basically the same expected values for our latent variable as obtained using the fitted function. Of course, the fitted function finds these estimates the right way, computing a prediction for each posterior sample rather than only using the posterior mean.
cor (predictions_latent[,1], mu)
## [1] 1
sd (predictions_latent[,1] - mu)
## [1] 3.571143e-16We can also ask for predictions as probabilities, rather than latent variable values, by not specifying the scale:
# predict probability of category membership
predictions_latent = fitted (model_ordinal, re_formula = NA)In this case our prediction results in a three dimensional matrix where response category varies along the third dimension (very much like our multinomial predictions). For example, below we see the probability of observing the first response category (small), for the first six observations.
# see first six for the first category
head (predictions_latent[,,1])
## Estimate Est.Error Q2.5 Q97.5
## [1,] 0.9557 0.016006 0.9184 0.9803
## [2,] 0.9314 0.026561 0.8714 0.9714
## [3,] 0.9799 0.009882 0.9566 0.9933
## [4,] 0.9763 0.008675 0.9561 0.9896
## [5,] 0.9435 0.017720 0.9035 0.9723
## [6,] 0.9941 0.003051 0.9866 0.9981If we consider only the first element of the second dimension (columns), we can get the expected value for each category, for the first six observations.
# see first six for each category
head (predictions_latent[,1,])
## P(Y = 1) P(Y = 2) P(Y = 3)
## [1,] 0.9557 0.043713 5.615e-04
## [2,] 0.9314 0.067657 8.960e-04
## [3,] 0.9799 0.019885 2.480e-04
## [4,] 0.9763 0.023454 2.943e-04
## [5,] 0.9435 0.055775 7.227e-04
## [6,] 0.9941 0.005824 7.232e-05We can use the expected value for the latent variable calculated manually above (mu), combined with the model thresholds, to calculate the expected probability that the observation will belong to each category (rounded to 5 decimal places).
predictions_manual =
cbind (round (plogis (threshold_1, mu),5),
round ((plogis (threshold_2, mu)-plogis (threshold_1, mu)),5),
round (1-plogis (threshold_2, mu), 5))
head (predictions_manual)
## [,1] [,2] [,3]
## [1,] 0.95834 0.04115 0.00051
## [2,] 0.93592 0.06328 0.00080
## [3,] 0.98201 0.01778 0.00021
## [4,] 0.97772 0.02202 0.00027
## [5,] 0.94604 0.05329 0.00067
## [6,] 0.99478 0.00515 0.00006Again, we can see that this approach results in basically the same values as those calculated with the fitted function.
cor (predictions_latent[,1,1],predictions_manual[,1])
## [1] 1
cor (predictions_latent[,1,2],predictions_manual[,2])
## [1] 1
cor (predictions_latent[,1,3],predictions_manual[,3])
## [1] 1Below we find the maximum value across each row of our predictions in predictions_manual, representing the hard, categorical prediction for each observation.
SG_hat = apply (predictions_latent[,1,],1,which.max)
SG_hat_manual = apply (predictions_manual,1,which.max)We can see that these two approaches lead to the same outcomes and that in either case we can correctly predict about 83% of our observations using only our fixed effects.
# manual and automatically-calculated predictions
mean (SG_hat == SG_hat_manual)
## [1] 1
# predictive accuracy for actual size group responses
mean (SG_hat == exp_data$SG)
## [1] 0.8264
mean (SG_hat_manual == exp_data$SG)
## [1] 0.8264Our focus has been on manually replicating some of the helper functions included in brms in order to highlight the relationship between our model parameters and expected outcomes. For more complex models, the specific effects of parameters or combinations of parameters can still be investigated using the techniques discussed in earlier chapters. For example, imagine an ordinal model involving a single quantitative predictor and a single categorical predictor with four levels. The geometry of the lines formed to predict expected values in such a model would be the same as that of the models discussed in Chapter 9, and many of the strategies discussed in that chapter could be directly applied to such an analysis.
12.3.5 Listener-specific discrimination terms
Before wrapping up our discussion of ordinal models, we want to talk briefly about the disc (discrimination) parameter. As mentioned above, this parameter is equal to the inverse of the scale parameter of the logistic distribution. In our previous ordinal model, the disc parameter was fixed at a value of 1 for all observations, However, this parameter can be predicted just as the error term (sigma, \(\sigma\)) of our normal or t-distributed errors was predicted in Chapter 8. Since this value cannot be negative our model will predict the logarithm of disc just as it did for sigma.
Below we fit a model with listener-specific discrimination parameters. We’re not going to discuss this in detail, however, the additional model structure is extremely similar to that presented in Chapter 8 (in particular Section 8.6.1), and should be clear based on the information provided in that chapter.
model_formula = bf(SG ~ vtl+f0 + (vtl+f0|L) + (1|S),
disc ~ 1 + (1|L))
priors =
c(set_prior("student_t(3, 0, 3)", class = "Intercept"),
set_prior("student_t(3, 0, 3)", class = "b"),
set_prior("student_t(3, 0, 3)", class = "sd"),
set_prior("student_t(3, 0, 1)", class = "Intercept",dpar="disc"),
set_prior("student_t(3, 0, 1)", class = "sd",dpar="disc"),
set_prior("lkj_corr_cholesky (2)", class = "cor"))
model_ordinal_disc =
brms::brm (model_formula, data=exp_data, family="cumulative", chains=4,
cores=4, warmup=1000, iter = 5000, thin = 4, prior = priors,
control = list(adapt_delta = 0.99))# Or download it from the GitHub page:
model_ordinal_disc = bmmb::get_model ("12_model_ordinal_disc.RDS")We can see that our model summary now includes a fixed effect for the disc_Intercept and the standard deviation of the listener-dependent discrimination terms (sd(disc_Intercept)) in the listener random effects section.
bmmb::short_summary (model_ordinal_disc)
## Formula: SG ~ vtl + f0 + (vtl + f0 | L) + (1 | S)
##
## Group-Level Effects:
## ~L (Number of levels: 15)
## Estimate Est.Error l-95% CI u-95% CI
## sd(Intercept) 0.95 0.42 0.36 1.99
## sd(vtl) 0.62 0.43 0.03 1.68
## sd(f0) 1.07 0.77 0.05 2.99
## sd(disc_Intercept) 0.42 0.11 0.25 0.67
## cor(Intercept,vtl) -0.19 0.36 -0.81 0.56
## cor(Intercept,f0) 0.21 0.37 -0.58 0.82
## cor(vtl,f0) -0.03 0.39 -0.76 0.72
##
## ~S (Number of levels: 139)
## Estimate Est.Error l-95% CI u-95% CI
## sd(Intercept) 2.43 0.85 1.11 4.33
##
## Population-Level Effects:
## Estimate Est.Error l-95% CI u-95% CI
## Intercept[1] -3.57 1.24 -6.36 -1.61
## Intercept[2] 4.97 1.72 2.30 8.83
## disc_Intercept -0.52 0.35 -1.17 0.21
## vtl 6.11 2.13 2.78 10.93
## f0 -3.31 1.51 -6.95 -0.95We can get the listener-dependent disc terms using the short_hypothesis function. Since these values are the logarithm of the discrimination, we need to exponentiate and then invert the parameter estimate to get the scale. We can do this inside the hypothesis function so that we summarize after manipulating (rather than exponentiating and inverting our summary).
listener_scale =
short_hypothesis (model_ordinal_disc,
"1/exp(disc_Intercept)=0", group="L",scope="coef")
# omit hypothesis column so this fits in the book
head(listener_scale)[,-5]
## Estimate Est.Error Q2.5 Q97.5 group
## H1 2.947 1.0599 1.3115 5.406 1
## H2 2.157 0.7857 0.9580 4.006 2
## H3 1.423 0.5277 0.6223 2.619 3
## H4 2.730 1.0239 1.1936 5.062 4
## H5 2.180 0.7832 0.9581 4.014 5
## H6 2.229 0.7954 0.9919 4.038 6Below we compare the distributions implied by these disc parameters for a value of 0 for the latent variable. We can see that these distributions imply substantially different behaviors across listeners, given the same predicted value for the latent variable.
show plot code
fixefs = fixef(model_ordinal_disc)
thresholds = fixefs[1:2,1]
xaxts = c('n','n','n','n','n',
'n','n','n','n','n',
's','s','s','s','s')
ymaxs = c(.2,.2,.2,.2,.2,
.2,.2,.2,.2,.2,
.35,.35,.35,.35,.35)
par (mfrow = c(3,5), mar = c(.15,.2,.2,.2), oma = c(4,3,1,1))
i=1
for (i in 1:15){
x = seq (-15,15,.1)
y = dlogis (x,0,listener_scale[i,1])
max_y = max(y)
y = y/max_y
plot (x, y, type = 'l',yaxt='n',xaxt=xaxts[i],
yaxs = 'i',xaxs='i',xaxt='n', ylim = c(0,1.4)) #ylim=c(0,ymaxs[i])
if (xaxts[i]=='s') axis(side=1,at = seq(-12,12,4))
x = seq (-15,thresholds[1],.1)
y = dlogis (x,0,listener_scale[i,1])
y = y/max_y
y = c(0,y,0)
x = c(x[1],x,x[length(x)])
polygon (x,y,col=bmmb::cols[1],border=bmmb::cols[1])
x = seq (thresholds[1],thresholds[2],.1)
y = dlogis (x,0,listener_scale[i,1])
y = y/max_y
y = c(0,y,0)
x = c(x[1],x,x[length(x)])
polygon (x,y,col=bmmb::cols[11],border=bmmb::cols[11])
x = seq (thresholds[2],15,.1)
y = dlogis (x,0,listener_scale[i,1])
y = y/max_y
y = c(0,y,0)
x = c(x[1],x,x[length(x)])
polygon (x,y,col=bmmb::cols[10],border=bmmb::cols[10])
x = seq (-15,15,.1)
y = dlogis (x,0,listener_scale[i,1])
y = y/max_y
lines (x, y, type = 'l',yaxt='n',xaxt=xaxts[i],lwd=2)
abline (v = thresholds,col=bmmb::cols[15], lwd=2)
p1 = plogis (thresholds[1],0,listener_scale[i,1])
p3 = 1 - plogis (thresholds[2],0,listener_scale[i,1])
p2 = (1-p3) - p1
text (-8,1.2,round(p1,2))
text (0,1.2,round(p2,2))
text (8,1.2,round(p3,2))
}
mtext (side=1,"Centered vocal-tract length (cm)", outer = TRUE, line = 2.9, cex = 0.9)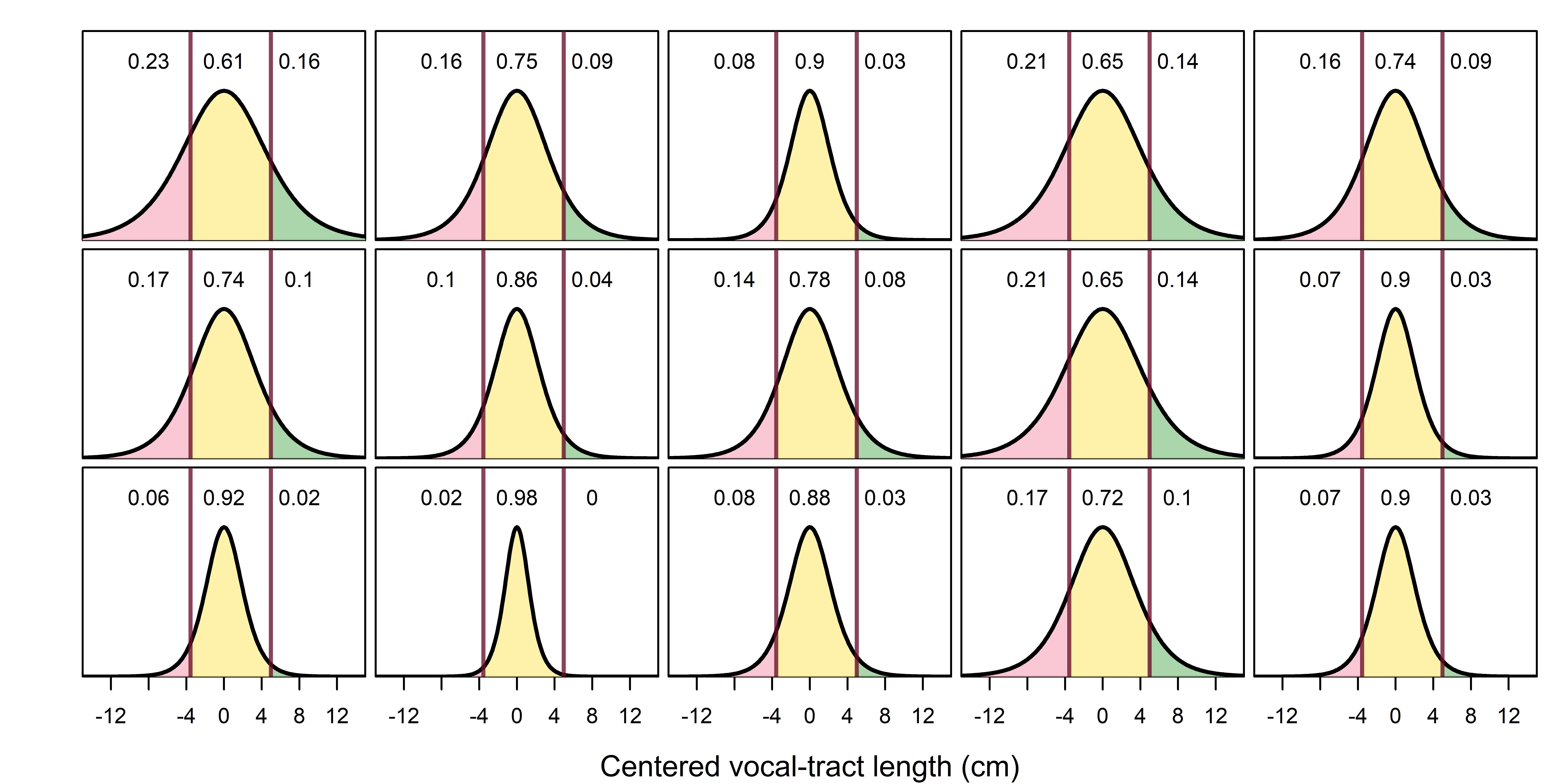
12.3.6 Answering our research questions
Our research question was very simple:
(Q1) Can we predict size group responses for a speaker given their f0 and VTL?
Yes we can, and with reasonable accuracy (83%) based only on two acoustic predictors. Below we find the predicted category for each trial based on our first model, including the random effects, and for the second ordinal model we fit that included listener-specific disc parameters.
# predictions with random effects
predictions_latent_full = fitted (model_ordinal)
# predictions with listener dependent disc
predictions_latent_disc = fitted (model_ordinal_disc)
# find highest predicted probability
SG_hat_full = apply (predictions_latent_full[,1,],1,which.max)
SG_hat_disc = apply (predictions_latent_disc[,1,],1,which.max)Below, we calculate the probability of a correct size group prediction for the fixed effects predictions, the predictions made by the full model with a single disc parameter, and the model with a listener-dependent disc parameter. We can see that the random effects noticeably improve prediction whereas the inclusion of listener-specific disc has a much smaller effect.
# fixed effects
mean (SG_hat == exp_data$SG)
## [1] 0.8264
# full model, single disc
mean (SG_hat_full == exp_data$SG)
## [1] 0.8887
# full model, listener-dependent disc
mean (SG_hat_disc == exp_data$SG)
## [1] 0.8911In any case, it seems clear that ordinal variables related to apparent talker size/height are likely to be very predictable based on speaker VTL and f0. However, we’re not going to dig into these results very much since this dependent variable was sort of ‘made up’, and our intention was mostly to provide practical examples of ordinal regression analyses that take advantage of the information in repeated measures data.
12.4 Exercises
The analyses in the main body of the text all involve only the unmodified ‘actual’ resonance level (in exp_data). Responses for the stimuli with the simulate ‘big’ resonance are reserved for exercises throughout. You can get the ‘big’ resonance in the exp_ex data frame, or all data in the exp_data_all data frame.
Fit and interpret one or more of the suggested models:
Easy: Analyze the (pre-fit) model that’s exactly like
model_multinomial, except using the data inexp_ex(bmmb::get_model("12_model_multinomial_ex.RDS")).Easy: Analyze the (pre-fit) model that’s exactly like
model_ordinal, except using the data inexp_ex(bmmb::get_model("12_model_ordinal_ex.RDS")).Medium: Model
SGusing a multinomial model and compare results, predictions, coefficient estimates, etc. withmodel_multinomial.Hard: Expand on either of the models fit in this chapter, either by adding duration as a quantitative predictor, or by including resonance and its interaction with the other predictors.
In any case, describe the model, present and explain the results, and include some figures.
12.5 References
Agresti, A. (2018). An introduction to categorical data analysis. John Wiley & Sons.
Agresti, A. (2003). Categorical data analysis. John Wiley & Sons.
Kruschke, J. (2014). Doing Bayesian data analysis: A tutorial with R, JAGS, and Stan.
