library (bmmb)
data (exp_data)
men = exp_data[exp_data$C_v=='m',]
# create vector of height judgments
mens_height = men$height4 Inspecting a ‘single group’ of observations using a Bayesian multilevel model
In the last chapter, we built a Bayesian regression model suitable for inspecting the average of a single group of observations. However, as we noted multiple times this model was ‘wrong’ for the structure of our data. The reason for this is that this model did not properly account for the repeated measures in our data. To properly handle repeated measures data, we need a multilevel model. In this chapter we will explain what we mean by ‘repeated measures’ and ‘multilevel’, in addition to fitting our first proper multilevel Bayesian model using brms.
4.1 Chapter pre-cap
In this chapter we discuss the analysis of data made up of multiple observations from members of a ‘single group’. We explain the ‘multilevel’ component of ‘Bayesian multilevel models’, and explain what is meant by ‘repeated measures data’. We introduce the problem of estimating factors with large numbers of levels, and present three approaches: No pooling, complete pooling, and adaptive partial pooling. After this, we fit a multilevel model with a structure that is more appropriate for our repeated measures data than the models presented in the previous chapter. We introduce the concepts of ‘random’ and ‘fixed’ effects and discuss how to inspect and manipulate random effects. We then simulate some repeated-measures data based on the parameters estimated by our model and see how the exclusion of different components affects our simulated data. Finally, we fit and interpret a model with a second ‘random’ effect to our data, and investigate the ‘shrinkage’ in the estimates of our ‘random’ model parameters.
4.2 Repeated measures data
Depending on the field of study, experiments often produce repeated measures data, data where multiple observations come from the same experimental unit (e.g. individual listeners, as in the data used in this book). As a practical matter, setting up experiments is often time-consuming such that it often makes more sense to collect, for example, 100 observations from each of 50 different people, rather than 1 observation from each of 5,000 different people. In addition, collecting more than one measurement from each source can go a long way toward reducing uncertainty in a model. However, the statistical analysis of repeated measures data requires models that take the repeated nature of the measurements into account. Treating repeated measures data as if it were not repeated measures data can cause problems for the inferences we make using statistical models.
For example, in our experiment, we are interested in the average apparent height of different speakers, or categories of speakers. Imagine we had an experiment with 10,000 total observations. Would it matter how these were distributed? What if we had 5,000 observations for each of two different speakers? In this case, we would have a lot of information about the apparent height of these speakers but not much information about speakers in general. What if we had 10 observations for each of 1,000 different speakers? In this case, we would have much more information about the apparent height of speakers in general, given the same number of overall observations. However, we have a lot less information about any individual speaker in the sample. So, we see two situations with the same total number of observations but a substantially different internal structures.
In general, the reason repeated measures data can cause problems when not modeled appropriately is because the observations within each experimental unit are not independent: Multiple measurements from the same person are probably going to be similar to each other. This can give us a warped perspective regarding the degree and nature of the variability in a set of observations when repeated measures data is not modeled appropriately. For example, the final model we used to analyze our data in the last chapter looked like this:
\[ \begin{equation} \begin{split} \\ height_{[i]} \sim \mathrm{N}(\mu_{[i]},\sigma) \\ \mu_{[i]} = \mathrm{Intercept} \\ \\ \textrm{Priors:} \\ \mathrm{Intercept} \sim \mathrm{N}(176, 15) \\ \sigma \sim \mathrm{N}(0, 15) \\ \end{split} \end{equation} \tag{4.1}\]
We’re going to consider the same data we discussed in Chapter 2 and Chapter 3, the apparent height judgments made for the adult male speakers in our experiment in the actual, unmodified resonance condition (see Section 1.3). Below we load the data that includes only these trials (exp_data) and subset to include only veridical male speakers:
Figure 4.1 presents the height judgments collected for adult male speakers in our experiment, organized in two different ways. In the left plot, we see the marginal distribution of the data with no differentiation made for the data collected from different listeners. This boxplot shows the individual observations of our vector \(height_{[i]}\) around the overall mean \(\mu\) with a standard distribution equal to \(\sigma\). In other words, the left boxplot represents the model presented in Equation 4.1. From the perspective of this model, an observation of 181 cm is +7 above the mean, regardless of who provided it, because 174 cm is the assumed mean for all listeners. As a consequence, all variation about the mean is noise. There is no systematic structure ‘inside’ that single giant box of data.
show plot code
################################################################################
library (bmmb)
data (exp_data)
men = exp_data[exp_data$C_v=='m',]
# create vector of height judgments
mens_height = men$height
#par (mar = c(.1,.1,.5,.1), mfrow = c(1,1), oma = c(4.2,.20,0,.2))
#layout (m=matrix(c(1,2,3,4),2,2), heights = c(.25,.75))
par (mar = c(4,.5,.5,.5), mfrow = c(1,2))
#plot (0,type='n',xlab='',ylab='',xaxt='n',yaxt='n',bty='n')
boxplot (men$height,col = 'grey', ylim = c(140,195),
width = 2, ylab="", horizontal = TRUE)
mtext (side=2,outer=TRUE, text="Apparent height (cm)", cex=1, line=2.9,adj=.55)
#boxplot (tapply(men$height,men$L,mean), xlab = "",xaxt='n',
# col = cols, ylim = c(140,195), width = 2,cex.axis=1.3,
# horizontal = TRUE)
#abline (h=)
#text (1,190,expression(sigma["[within]"]), cex=2)
boxplot (height ~ L, data = men, xlab = "", ylab="",xaxt='s',
col = cols[-1], ylim = c(140,195), cex.lab=1.1,cex.axis=1.1,
horizontal = TRUE, yaxt='n')
grid()
abline (h = mean(mens_height), lwd=2, lty=3)
boxplot (height ~ L, data = men, col = cols[-c(1,8)],add=TRUE,yaxt='n',xaxt='n')
mtext (side=1,outer=TRUE, "Apparent height (cm)",line=-0.9)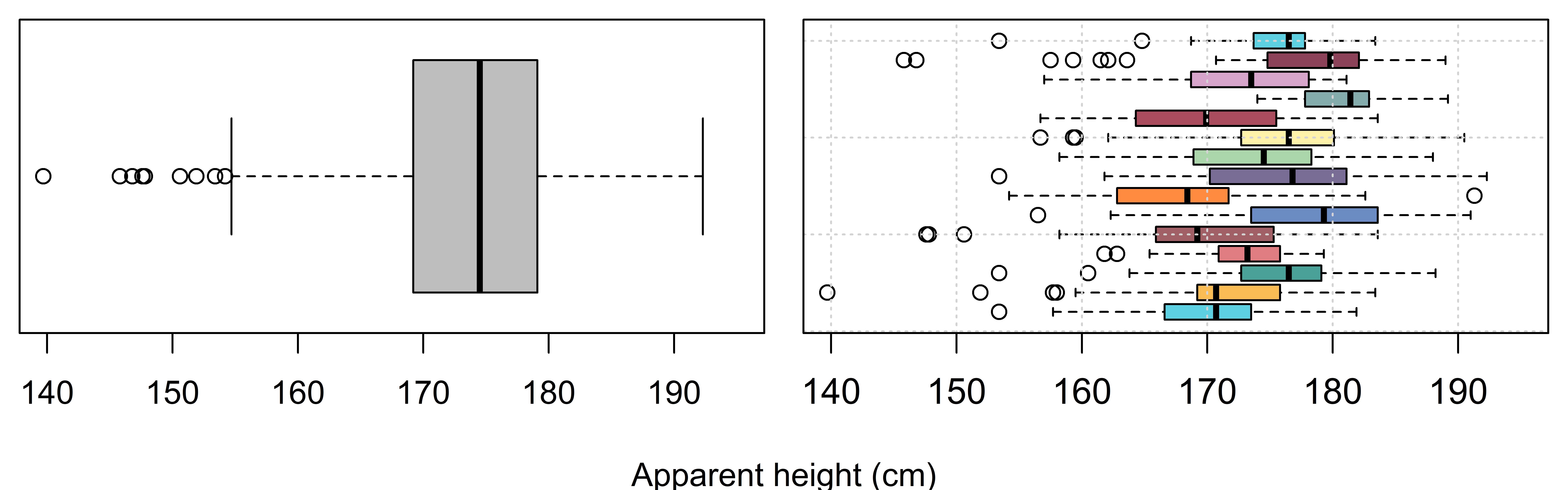
In the right plot, we see each listener’s height judgments in a different boxplot. Clearly, each listener has a general tendency so that certain values are more or less surprising for individual listeners. For example, a height response of 181 cm is perfectly average for listener 6 (blue box, sixth from the bottom). Thus, if you know listener 6 provided a response, an apparent height of 181 cm should not be surprising. In fact, this would represent an error of 0 cm when provided by this listener, considered relative to their expected apparent height judgment. In contrast, a response of 174 cm, the overall average response, would be a relatively atypical response for this listener.
The failure to acknowledge the fact that listeners provided non-independent responses might not seem like a big deal but it really is. Remember from Section 2.3.2 that the calculation of joint probabilities (and densities) is ‘easy’ only if the model residuals are assumed to be independent. Well, on the right side of Figure 4.1 we see that all of our errors (i.e. deviations from the intercept at 174 cm) are definitely not independent, which means that ignoring this fact is going to lead to inaccuracies throughout our models. It’s also a fact that we actually do have different listeners providing our data, and they do appear to vary from each other, and maybe we want to represent this in our model.
4.2.1 Multilevel models and ‘levels’ of variation
We can think of the repeated measures data presented in Figure 4.1 as having multiple ‘levels’ of variation. These levels are often referred to as upper and lower but this is entirely metaphorical/conceptual. Another way to think of them is as within-group variation (lower) and between-group variation (upper). We will describe these ‘levels’ of variation with respect to our repeated measures data involving the apparent heights of adult males presented in Figure 4.1.
The ‘lower’ level: The data-level distribution of individual observations given a certain listener. This is the distribution of data points inside of each of the little boxes in the right plot in Figure 4.1. If not for this variation, the boxes in the plot would all be clustered about their respective listener means. So, the lower level of variation gives the listener boxes their widths, and causes one response provided by a listener to differ randomly from another response from the same listener. For the sake of simplicity, our models usually assume that the standard deviation is equal to \(\sigma\) for all of the listener-dependent distributions, though our models can relax this assumption (see Chapter 8).
The ‘upper’ level: The distribution of parameters between your sources of data. The upper level of variation causes the boxes in the right plot of Figure 4.1 to not all be aligned at the grand mean. Although the listener mean is a parameter for the distribution of apparent height responses given the listener, it can also be thought of as a random variable. When we run an experiment we randomly ‘sample’ a small number of values of this variable by selecting a small number of humans to participate in our experiment. We might expect the average reported height for a single listener to be a relatively stable property such that a replication with the same listener should result in about the same listener mean. However, it’s impossible to know any given listener’s average apparent height judgment a priori. As a result, our listener means are as unpredictable to us as the individual responses provided by those listeners.
A multilevel model is a special kind of regression model that’s able to simultaneously estimate random variation at multiple ‘levels’. A multilevel model fit to our height judgments would be able to estimate the average reported by each listener, the variation in listener means, and the random within-listener variation, all at the same time. Model parameters describe some tendencies in the data. For now we’re just talking about average values. So, our model describes listeners in terms of their average response, which it encodes using parameters related to the listener means. As a result, when we say “the distribution of parameters” to refer to the distribution of listener means, we’re really saying “the distribution of listener characteristics”. So, when we say “multilevel models model variation at the data level and the parameter level”, we’re basically saying “multilevel models allow us to model the variation of data within listeners, and of parameters/characteristics between listeners”.
Figure 4.2 compares visual representations of the same data, the points \(y_{[1]} \dots y_{[6]}\), represented as unilevel or multilevel models. In the unilevel model there is only one distribution, the data distribution, and a single random source of variation, the data-level error \(\sigma\). Since there is no ‘upper’ level describing between-listener variation, our model doesn’t ‘know’ our data contains repeated observations from a small number of listeners. In addition, our model has no mechanism by which to explain, for example, why \(y_{[1]}\) and \(y_{[2]}\) have such low values. This model treats all deviations from the mean as random and therefore acts as if systematic between-listener variation did not exist. It also treats all variation around the mean as totally independent, which we know it is not.
show plot code
################################################################################
knitr::include_graphics("images/Figure 4.2.jpg")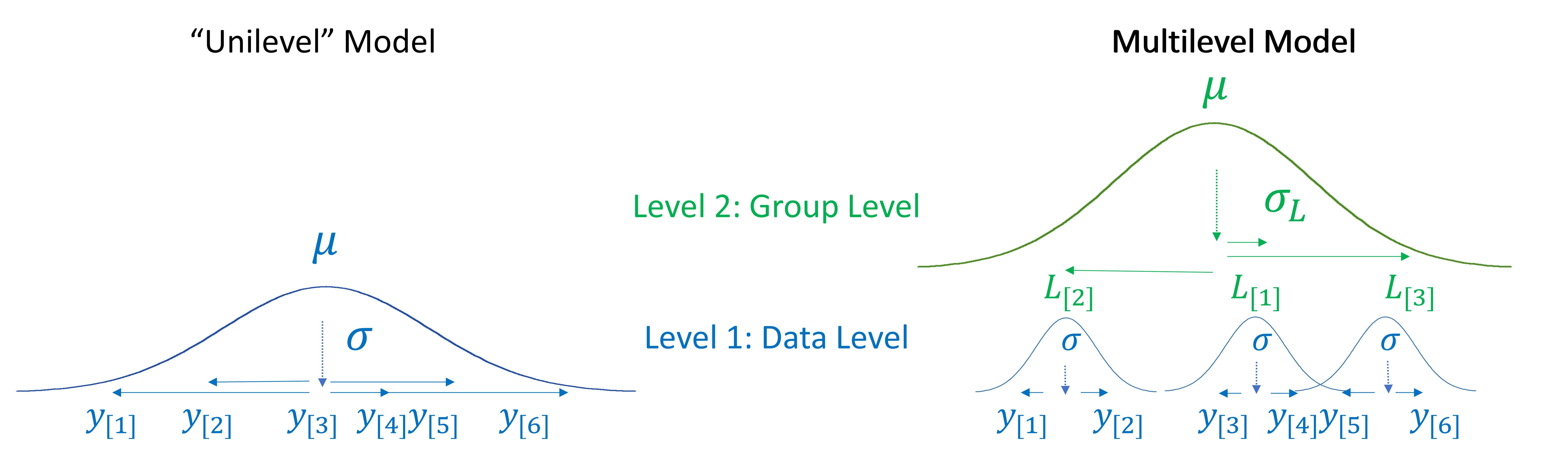
In contrast, the multilevel model in the same figure introduces an intervening second level between the observations and the grand mean \(\mu\). This level features some new symbols, these are:
listener-specific effects representing the difference between the listener mean and the grand mean, \(L_{[j]}\) for listener j.
\(\sigma_L\), the standard deviation of the listener effects.
Notice that we are now estimating two variance components, the data-level error (\(\sigma\)) and the random between-listener variation \(\sigma_L\). This seemingly small difference in structure results in important changes in our model. For example, since the total amount of variation in the data is fixed, as variation in the second level (\(\sigma_L\)) grows larger, variation at the first level (\(\sigma\)) necessarily gets smaller.
We can see this happening in Figure 4.2. In the unilevel model, \(y_{[1]}\) and \(y_{[2]}\) where low values ‘for no reason’, resulting in large deviations from our expectations and a large \(\sigma\) (the blue distribution). Now, we know that \(y_{[1]}\) and \(y_{[2]}\) are low because listener 2 (\(L_{[2]}\)) tends to provide low values. As a result, the large deviations between \(y_{[1]}\), \(y_{[2]}\), and the grand mean (\(\mu\)) are mainly attributed to \(\sigma_L\) (the green distribution) rather than \(\sigma\). In the multilevel model, it is only the relatively small amount of within-listener variation that is directly attributed to \(\sigma\), resulting in a substantial reduction in the data-level error.
One thing to note is that if there is no between-listener variation, i.e. if \(\sigma_L=0\), the the multilevel model above converges structurally on the unilevel model. For example, if \(\sigma_L\) is near zero all the listener effects would need to have very small values. If this occurred, all the densities at the lower level in the right plot in Figure 4.2 would overlap, and look a lot like the single density in the left side of the figure. In addition, the second level distribution would effectively be meaningless/useless since it would represent no systematic variation between listeners. This potential equivalence is a very important feature of multilevel models. Since the multilevel model estimates values of \(L\) and \(\sigma_L\), the outcome \(\sigma_L \approx 0\) is within the realm of possibility. So, fitting a multilevel model allows one to investigate the very existence of systematic variation in parameters across research units (e.g. listeners) but does not necessarily impose this on data if such systematic variation is not present.
4.3 Representing predictors with many levels
Before moving on to a discussion of multilevel models and parameter estimation strategies, we want to take a moment to discuss some practical and notational issues that arise when your models include factors with large numbers of levels. First, we will discuss a way to efficiently represent models including factors with many levels as predictors. After that, we will discuss some ‘quirks’ in the way that R names model parameters and the way that we’ve chosen to represent this in our model notation.
R treats nominal, categorical predictors (such as listener) as factors and assumes that each different label represents a different group. Each value of a factor is called a level. Although we can indicate listeners using numbers, we’re not treating this predictor as numerical or otherwise quantitative. Instead, we treat listener (L) as a factor, and the individual listeners in the experiments are its levels. As far as our models are concerned, participant/speaker/subject/listener has no special status as a predictor and it is just a factor with (usually) many levels.
In order to estimate a separate effect for each listener, we need to add 15 parameters to our model, \(L_{[1]},...,L_{[15]}\), one for each listener. In order to represent these predictors, our model descriptions could include a predictor for each listener coefficient that would equal one or zero based on whether the row was contributed by that listener (1) or not (0). For example, in Equation 4.2 we see a line consisting of a set of 15 predictors associated with the \(L\) coefficients, all multiplied by zero save for the second one. The effect of this, seen in the second line in Equation 4.2, is to only have the second listener coefficient contribute to the predicted value for observation \(i\). In other words, observation \(i\) was contributed by listener 2 for this data.
\[ \begin{equation} \begin{split} \mu_{[i]} = \mathrm{Intercept} + L_{[1]} \cdot 0 + L_{[2]} \cdot 1 + L_{[3]} \cdot 0 +\ldots + L_{[15]} \cdot 0 \\ \mu_{[i]} = \mathrm{Intercept} + L_{[2]} \\ \end{split} \end{equation} \tag{4.2}\]
This is actually how regression models represent these sorts of predictors ‘under the hood’. However, if every single factor level were to get an independent parameter represented in the regression equations in this book, these would become very long and difficult to interpret. For example, we have 139 different speakers in our data, meaning our prediction equation would be extremely long if we included each of these predictors as separate terms. Instead, we can treat the effects associated with the levels of a factor as a vector, and just pick the correct element of this vector for each observation in our data. In order to know which listener contributed each observation, we also need an index variable, a variable that equals a number from one to 15 that indicates which listener parameter to use on that trial. This is the L column in our data (exp_data), the listener predictor.
For example, below we see that our listener predictor has a value of 1 for observation 1 and a value of 8 for observation 1,000. This tells us that the first listener provided the first observation and the eighth listener provided the 1000th observation. It tells our model to use \(L_{[1]}\) for observation 1 and \(L_{[8]}\) for observation 1,000. In general, we should use \(L_{[j]}\) for observation \(i\) when the listener prediction is equal to \(j\) for that observation (i.e. exp_data$L[i]=j).
exp_data$L[1]
## [1] 1
exp_data$L[1000]
## [1] 8Unfortunately, this leads to some notational strangeness. R names the vector of coefficients in your model using the same name as your predictor. So, the vector of listener effects in your model won’t have an interpretable name like listener_effects. Instead, we have a predictor called L which represents listeners, and, we have a set of coefficients called \(L\) in our model that represent the effects for each of our listeners. As a result, our regression equations will include terms that look like this, \(L_{[\mathsf{L}_{[i]}]}\), which means: “Coefficient \(L\), and the level is the one indicated by value of the L predictor (\(\mathsf{L}\)) for trial \(i\)”. This term is now included in our prediction equation in Equation 4.3, which says: The expected value for any given trial is equal to the intercept, plus the listener effect (\(L\)) specific to the listener that contributed that trial as indicated by the L vector (\(\mathsf{L}\) in our model).
\[ \begin{equation} \begin{split} \mu_{[i]} = \mathrm{Intercept} + L_{[\mathsf{L}_{[i]}]} \end{split} \end{equation} \tag{4.3}\]
We think the \(L_{[\mathsf{L}_{[i]}]}\) notation is potentially confusing, and not ideal. However, the only way we can have formal model descriptions that actually match the name of our model coefficients and the names of the predictors they are associated with is by giving these the same names. One thing we’ll do to try to make things clearer is use different font for the predictors (\(\mathsf{L}\)) and the corresponding coefficients (\(L\)). So, when you see terms like these \(L_{[\mathsf{L}_{[i]}]}\) in your models, this is nothing more than a compact way of representing the same model structure shown in Equation 4.2.
For example, if your data contains a predictor called L \((\mathsf{L})\) whose first three values are 2, 4, and 1, then we need parameters \(L_{[2]}\), \(L_{[4]}\), and \(L_{[1]}\) for the first three trials. We can get the correct listener effect for each trial using the information in our listener predictor. We can see how this would work in the example in Equation 4.4, showing the combination of intercepts and listener effects for the first three observations in our hypothetical example. In each case, we find the value of the predictor \(\mathsf{L}\) for observation \(i\) and use this to fetch the correct listener effect (\(L\)) for our regression equation for that observation.
\[ \begin{equation} \begin{split} \mathsf{L}_{[1]} = 2, \mathsf{L}_{[2]}=4,\mathsf{L}_{[3]}=1, \dots \\ \\ \mu_{[i=1]} = \mathrm{Intercept} + L_{[\mathsf{L}_{[i=1]}]} = \mathrm{Intercept} + L_{[2]} \\ \mu_{[i=2]} = \mathrm{Intercept} + L_{[\mathsf{L}_{[i=2]}]} = \mathrm{Intercept} + L_{[4]} \\ \mu_{[i=3]} = \mathrm{Intercept} + L_{[\mathsf{L}_{[i=3]}]} = \mathrm{Intercept} + L_{[1]} \\ \dots \end{split} \end{equation} \tag{4.4}\]
The example above says: We add the intercept to the effect for listener 2 to get our expected value for observation 1 (i.e. observation 1 was contributed by listener 2), we add the intercept to the effect for listener 4 to get our expected value for observation 2 (i.e. observation 2 was contributed by listener 4), and so on, for all observations in our data.
4.4 Strategies for estimating factors with many levels
We know that the 15 values of \(L\) are related by being different listeners in our experiment. The way you tell your model that these coefficients are related is by treating them as levels of the same factor rather than as unrelated coefficients. One way to think of factors is that they represent ‘batches’ of coefficients in your model that are thematically or conceptually related (more on this in chapter 11). Once you encode the fact that there are ‘batches’ of parameters in your model, you have several options with respect to how these batches are estimated.
Gelman and colleagues (Gelman and Hill, 2006; Gelman et al. 2013) distinguish three general ways that parameters related to levels of a factor can be estimated: (1) Complete pooling, (2) No pooling, and (3) Partial pooling. The distinction between complete, no, and partial pooling is made for each factor in a model. So, one factor may be estimated using partial pooling while another may be estimated using no pooling. This makes understanding the distinction between these approaches very important.
4.4.1 Complete pooling
In Chapter 3, we carried out a complete pooling analysis of our data, presented in Equation 4.1. That is, we threw everything into one big pile and analyzed it as if it came from one undifferentiated group (as in the unilevel model in Figure 4.2). There are several problems with this approach when applied to our experimental data. First, the complete pooling approach does not allow us to make any statements about variation between listeners since it does not estimate values of \(L\). In fact, this model assumes that \(\sigma_L\) is equal to zero (or does not exist). As noted earlier, if listeners do not differ from each other in any way, then \(\sigma_L\) will be equal to zero and the two models in Figure 4.2 will converge on each other. However, if \(\sigma_L\) is not zero, then this approach will miss out on important information.
There is perhaps a more serious problem with the complete pooling approach for repeated measures data. The complete pooling model assumes that all deviations from the mean are due to random error. Recall that regression models assume that all random variation around expected values (i.e. the residuals) are independent. Since independence is assumed, likelihoods are calculated the ‘easy’ way by multiplying individual marginal probabilities (see sections Section 2.3.2 and Section 2.7.1). However, as we can see in Figure 4.1, each listener had a slightly different average judged height. This means that the residuals associated with each individual listener are not independent and may be related. For example, we expect almost all of listener 12’s residuals to be positive (i.e. larger than average) since nearly all of this listener’s responses were above the overall average \(\mu\). The violation of the assumption of independent residuals means that if we’re using a complete pooling approach for our repeated measures data we are calculating likelihoods the ‘wrong’ way. Basically, our data has 675 rows but not 675 totally independent observations, but our model doesn’t know this. As a result, the information provided by our model may not be reliable.
We’ve previously mentioned that our models are not exactly ‘right’ or ‘true’, so why does it matter all of a sudden that the complete pooling model is ‘wrong’? Recall that in Section 2.7.1 we mentioned that the likelihood function gets narrower as a function of the sample size n. This applies only to n independent observations, but not necessarily to dependent observations. As a result, when we treat dependent observations as independent our likelihood may end up being narrower than is warranted. This gives us a false sense of security about how precise our parameter estimates are. Basically, spherical models of billiards balls are wrong but useful, while cubic models of the balls are wrong and less useful. Treating residuals both within and across listeners as independent is kind of like using a cubic model of a billiard ball.
4.4.2 No pooling
To account for systematic variation between listeners in the data we can include listener predictors (\(L\)) in our model, as in Equation 4.5. When we estimate the levels of this factor with no pooling, we estimate every level of the factor totally independently. One way to model this mathematically is to think of these coefficients as coming from a uniform distribution that extends from negative to positive infinity. The uniform distribution is just a flat line, meaning any value in the interval is equally likely. So, a uniform distribution extending from positive to negative infinity places absolutely no restrictions on the possible values of a parameter and says absolutely any value is equally likely. This is equivalent to setting \(\sigma_L=\infty\). As a result, we can see complete and no pooling as two extremes, in one case we assume there is no between-listener variation at all and in the other we assume that between-listener variation can potentially be infinitely large.
In Equation 4.5 we see that in our no pooling model, the expected value (\(\mu\)) for each trial is the sum of our intercept and the listener predictor, and that each of the \(L\) terms is drawn from the same uniform distribution. We introduce the notation \([\bullet]\) to represent every individual level of a factor. Below, we note that each of the 15 listener effects (\(L_{[\bullet]}\)) was individually drawn from the same uniform distribution.
\[ \begin{equation} \begin{split} height_{[i]} \sim \mathrm{N}(\mu_{[i]},\sigma) \\ \mu_{[i]} = \mathrm{Intercept} + L_{[\mathsf{L}_{[i]}]} \\ \\ \textrm{Priors:} \\ L_{[\bullet]} \sim \mathrm{uniform}(-\infty, \infty) \\\\ \mathrm{Intercept} \sim \mathrm{N}(176, 15) \\ \end{split} \end{equation} \tag{4.5}\]
There are two main weaknesses to this approach. First, assuming that \(\sigma_L=\infty\) does not allow regularization (also known as shrinkage, see Section 3.3.3) to occur across the levels of the factor. Shrinkage occurs when an estimate is pulled toward its prior mean. However, a perfectly uniform prior does not exert any influence on posterior distributions, so that these are completely reflective of the likelihood function. In the absence of shrinkage, the characteristics of each listener are estimated as if only a single listener had taken part in our experiment. But we don’t have one listener, we have 15, and they all performed the experiment in quite similar ways. Clearly, it may be useful if our model could consider what it knows about the values of \(L\) in general when it estimates the value of \(L_{[j]}\) for any given listener \(j\).
The second weakness of a no pooling approach is that these models do not represent information about the range of credible values for the levels of a factor. For example, below we see some quantiles for the average apparent height reported for adult male speakers by the listeners in our experiment:
listener_means = tapply (men$height, men$L, mean)
quantile(listener_means)
## 0% 25% 50% 75% 100%
## 167.8 170.5 174.1 175.9 180.9These listeners each carried out this experiment independently and did not know each other. Despite this, we see below that half of listeners’ average height judgments are between 171 and 176 cm, and that all listener averages fell between 168 and 181 cm. The consistency of these judgments suggests a common behavior between listeners, and an underlying similarity in listener characteristics and parameters. Given this, it might be surprising if a new listener carried out the experiment and provided an average height judgment of 130 cm for adult males. Although our model does include information about individual listeners, it cannot represent information about more or less likely values for listener effects.
4.4.3 (Adaptive) Partial pooling
Complete pooling and no pooling offer extremes: We either assume \(\sigma_L\) is zero or we assume it is infinite. Partial pooling offers a sort of middle ground between complete pooling and no pooling, a potential ‘just right’ compromise solution. Partial pooling estimates the values of \(L\) using non-zero values of \(\sigma_L\), meaning that the parameters representing the levels of a factor are neither completely independent nor totally merged. This allows the parameter estimates to share information and often results in the parameters being more similar to each other compared to when they are fit using no pooling.
Technically speaking, we can fit a model with partial pooling by specifying any finite prior probability for our parameters. So, simply drawing the \(L\) parameters from any given shared distribution (e.g. \(L_{[\bullet]} \sim \mathrm{N}(0,30)\)) would constitute a partial pooling estimate of the parameters. However, we know that our prior probability can help to pull parameters toward the prior mean (as discussed in Section 3.3.2), resulting in shrinkage. In order for this to work effectively, the variance of the prior needs to be set at a reasonable value given the amount of variation in the data. Below, we see that the standard deviation of listener means is 3.6 cm. It would be nice to be able to use a value close to this, since we know this to be a good fit for our data. Unfortunately, if we did this it would not really be a prior probability anymore, since we would be analyzing the data, finding the most likely parameter values, and then using that to set what is supposed to be the prior probability.
sd(listener_means)
## [1] 3.594Instead of doing this, we can estimate \(\sigma_L\) using our data. This allows us to have a prior distribution for our parameters that is tailored to the data, without us having to actually specify it a priori. When standard deviation parameters like \(\sigma_L\) are estimated from the data rather than specified a priori, this is called adaptive partial pooling. Often, the ‘adaptive’ part is dropped and authors may describe the method as partial pooling when they mean specifically adaptive partial pooling. So, when you read that a factor was estimated using ‘partial pooling’, most often that means that the standard deviation parameter corresponding to that factor (e.g. \(\sigma_L\) for \(L\)) was estimated from the data. We will refer to it as adaptive pooling, tending to drop the ‘partial’, in order to clearly communicate when we refer to parameters whose variance parameters (e.g. \(\sigma_L\)) are estimated from the data.
The adaptive (partial) pooling version of our model is presented in Equation 4.6. There are two important differences with respect to the model in Equation 4.5. First, the \(L_{[\bullet]}\) terms are drawn from a normal distribution with a mean of zero and a standard deviation equal to \(\sigma_L\). The reason the mean of the distribution is equal to zero is that our listener coefficients only encode deviations relative to the intercept, and not the listener average. So, a listener that is perfectly average will have a coefficient equal to 0. Second, since the \(\sigma_L\) term is estimated from the data a prior distribution for \(\sigma_L\) is specified. This is because prior probabilities must be specified for all estimated parameters in our model, and \(\sigma_L\) is now being estimated.
\[ \begin{equation} \begin{split} height_{[i]} \sim \mathrm{N}(\mu_{[i]},\sigma) \\ \mu_{[i]} = \mathrm{Intercept} + L_{[\mathsf{L}_{[i]}]} \\ \\ \mathrm{Priors:} \\ L_{[\bullet]} \sim \mathrm{N}(0,\sigma_L) \\ \\ \mathrm{Intercept} \sim \mathrm{N}(176,15) \\ \sigma \sim \mathrm{N}(0,15) \\ \sigma_L \sim \mathrm{N}(0,15) \end{split} \end{equation} \tag{4.6}\]
This approach is called adaptive partial pooling because the estimate of each level of \(L\) is partially influenced by every other level of \(L\) by virtue of being drawn from the same distribution \(\mathrm{N}(0,\sigma_L)\). For example, we saw that half of listener mean height judgments were between 170 and 176 cm, and our model will now take that into account when estimating the individual coefficients that make up \(L\). So, unlike the complete pooling case, we do not ignore variation between listeners, and unlike in the no-pooling case, we do not ignore the similarities between them. In addition, the amount of blending that occurs across levels is determined by the data, and so the method is ‘adaptive’ to the specific situation.
4.4.4 Hyperpriors
You may have noticed that in Equation 4.6 we assign a prior distribution to \(\sigma_L\). However, \(\sigma_L\) is a parameter in the prior distribution of our listener parameters \(L_{[\bullet]}\). So actually, the prior for \(\sigma_L\) is the prior of a prior. We might call this a ‘grand prior’ (like a grandparent) but the actual term for it is hyperprior. One way to think of it is that in a multilevel model, each level effectively acts as the prior for the level below it. This leads to a nested or hierarchical model structure (sometimes multilevel models are called hierarchical models for this reason).
We’re going to update the equation defining our posterior distributions (originally presented in Equation 3.12) to take into account the structure of our partial pooling models. Our model will now reflect not only our prior beliefs but also the information encoded in our hyperpriors, and it will estimate the distribution of \(\sigma_L\) from the data. We can begin with the posterior probability of observing a given value of \(L_{[\bullet]}\) for a single listener in isolation. Equation Equation 4.7 says that the posterior probability of some listener effect (\(L_{[\bullet]}\)) given the data \(y\) is equal to the product of the likelihood of \(L_{[\bullet]}\) given the data (\(P(y|L_{[\bullet]})\)) and the prior of \(L_{[\bullet]}\) (\(P(L_{[\bullet]})\)), divided by the marginal probability of the data (\(P(y)\)).
\[ \begin{equation} \begin{split} P(L_{[\bullet]}|y) = \frac{P(y|L_{[\bullet]}) \cdot P(L_{[\bullet]})}{P(y)} \end{split} \end{equation} \tag{4.7}\]
If we want to consider the posterior probability of \(\sigma_L\) given the data, we can add \(\sigma_L\) to every term that previously included \(L_{[\bullet]}\) in Equation 4.7. Basically, we’re saying \(\sigma_L\) and \(L_{[\bullet]}\), instead of just \(L_{[\bullet]}\). Equation Equation 4.8 can be read aloud as “the posterior probability of observing a certain listener effect and a certain amount of between-listener variation given some data (\(P(L_{[\bullet]},\sigma_L|y)\)) is equal to the likelihood of the data given the listener effect and the amount of variation between listeners (\(P(y|L_{[\bullet]} , \sigma_L)\)), times the joint prior probability of the listener effect and the amount of variation between listeners (\(P(L_{[\bullet]},\sigma_L)\)), divided by the marginal probability of the data (\(P(y)\))”.
\[ \begin{equation} \begin{split} P(L_{[\bullet]},\sigma_L|y) = \frac{P(y|L_{[\bullet]} , \sigma_L) \cdot P(L_{[\bullet]},\sigma_L)}{P(y)} \end{split} \end{equation} \tag{4.8}\]
We know that we can represent the joint probability of a listener effect and the listener standard deviation, \(P(L_{[\bullet]},\sigma_L)\), using normalized conditional probabilities as in \(P(L_{[\bullet]}|\sigma_L) \cdot P(\sigma_L)\) (see Chapter 2). We can also remove the listener standard deviation (\(\sigma_L\)) from the likelihood of \(L_{[\bullet]}\) given the data (\(P(y|L_{[\bullet]})\)), because we know that the probability of any given data point in a normal distribution depends only on the mean of the distribution and not the standard deviation of means between distributions (i.e. \(\sigma_L\)). These two changes are reflected in Equation 4.9. The first term in the numerator on the right-hand side is the likelihood, this was in our original equation in Equation 4.7. The second term is the prior distribution of listener effects. This also appeared in Equation 4.7, however, this distribution now allows for variation in the value of \(\sigma_L\). Finally, the third term (\(P(\sigma_L)\)) represents the hyperprior, the prior distribution of one of the parameters of another prior distribution.
\[ \begin{equation} \begin{split} P(L_{[\bullet]},\sigma_L|y) = \frac{P(y|L_{[\bullet]}) \cdot P(L_{[\bullet]}|\sigma_L) \cdot P(\sigma_L)}{P(y)} \end{split} \end{equation} \tag{4.9}\]
Our posterior estimates of \(L_{[\bullet]}\) may differ from our no pooling estimates of the listener effects. This is because, in addition to the individual parameter likelihoods, the posterior probability of \(L_{[\bullet]}\) in Equation 4.9 takes into account the estimates of \(\sigma_L\). When our posterior estimates of the listener (or any other) effects are more similar to the population average, and to each other, than the no pooling estimates, regularization has occurred. Importantly, the amount of pooling/shrinkage that will occur will be determined by \(\sigma_L\), which is estimated from the data itself.
4.5 Estimating a multilevel model with brms
We’re going to fit a model to the same data we investigated in Chapter 3, however, we’re going to use a multilevel model that reflects the repeated measures nature of the data.
4.5.1 Data and research questions
Below we load and subset our full data to only include those trials involving adult male speakers, and only when the actual resonance size was presented. This is the same data we considered in chapters 2 and 3.
# load book package and brms
library (bmmb)
library (brms)
## Warning: package 'brms' was built under R version 4.3.2
## Loading required package: Rcpp
## Loading 'brms' package (version 2.20.4). Useful instructions
## can be found by typing help('brms'). A more detailed introduction
## to the package is available through vignette('brms_overview').
##
## Attaching package: 'brms'
## The following objects are masked from 'package:bmmb':
##
## get_prior, prior_summary
## The following object is masked from 'package:stats':
##
## ar
# load and subset experimental data
data (exp_data)
men = exp_data[exp_data$C_v=='m',]Once again, we’re going to try to address the following basic research questions:
(Q1) How tall does the average adult male sound?
(Q2) Can we set limits on credible average apparent heights based on the data we collected?
To analyze our data using brms in R we need it to be in a data frame with each row containing a different observation. We need one column representing our dependent variable (the variable we want to analyze) and one column that indicates the ‘source’ of the data. The relevant columns in our data are:
L: An integer from 1 to 15 indicating which listener responded to the trial.height: A floating-point number representing the height (in centimeters) reported for the speaker on each trial.
We can see that our data satisfies these conditions below, where the relevant columns (for this model) are L for listeners and height for our dependent variable.
head (men)
## L C height R S C_v vtl f0 dur G A G_v A_v
## 93 1 m 169.9 a 47 m 14.8 172 339 m a m a
## 95 1 m 173.5 a 48 m 15.6 108 236 m a m a
## 97 1 m 172.0 a 49 m 15.5 96 315 m a m a
## 99 1 b 157.7 a 50 m 14.5 134 240 m c m a
## 101 1 m 153.4 a 51 m 15.0 122 241 m a m a
## 103 1 m 174.2 a 52 m 14.8 101 259 m a m aOften, experiments (or data more generally) will feature a single factor that indicated the ‘source’ of information. However, in this experiment, we have both speakers producing our data and listeners providing judgments about our data. This leads to non-independent observations based on both speaker and listener. For example, a speaker who sounded tall to one person may have sounded tall to other listeners, and listeners who identified all speakers as generally tall/short would have done so in a similar way across all speakers. We’ll begin by building a model that only incorporates information about listeners before discussing a model that includes information regarding both speakers and listeners later in the chapter.
4.5.2 Description of the model
To specify a multilevel model, you need to write a slightly more complicated model formula. This explanation assumes that you have a data frame where one column contains the variable you are interested in predicting (in this case height), and another column contains a vector containing unique labels for each grouping factor (e.g., speaker, listener, and participant) in this case a unique listener label L. Before, the model formula looked like this:
height ~ 1
Which meant ‘predict height using only an overall intercept’. To indicate that your model contains an ‘upper’ level where you have repeated measures clusters of data coming from different sources, you have to put another model formula inside your main model formula. The formula corresponding to the model shown in Equation 4.6 looks like this:
height ~ 1 + ( 1 | L)
Which means ‘predict height using an overall intercept and separate intercepts for each level of the L (listener) factor’. In mathematical notation, this symbol | (called ‘pipe’) means “given that” or “conditional on”. It can effectively be interpreted in the same way in our models: “estimate an intercept given each level of the L predictor”. So, when you place a predictor in parenthesis on the right-hand side of a pipe |, like this ( | predictor ), you tell brm that you would like to estimate separate parameters for each level of the factor. Whatever you put in the left-hand side of the parentheses ( in here | predictor ) specifies the parameters you wish to estimate for each level of the factor. In addition, any parameters specified on the left-hand side of the pipes in parentheses, ( in here | predictor ), will be estimated using adaptive partial pooling. By convention, effects estimated using adaptive pooling are modeled as coming from a normal distribution with a mean of zero, so that aspect of the model does not need to be directly specified.
So, the model formula height ~ 1 + ( 1 | L) tells brms to build an intercept-only model with a separate intercept for each listener, and that the listener intercept terms should be estimated with adaptive pooling. This model formula specifies a subset of the information provided in our full model specification shown in Equation 4.10 (originally Equation 4.6).
\[ \begin{equation} \begin{split} height_{[i]} \sim \mathrm{N}(\mu_{[i]},\sigma) \\ \mu_{[i]} = \mathrm{Intercept} + L_{[\mathsf{L}_{[i]}]} \\ \\ \mathrm{Priors:} \\ L_{[\bullet]} \sim \mathrm{N}(0,\sigma_L) \\ \\ \mathrm{Intercept} \sim \mathrm{N}(176,15) \\ \sigma \sim \mathrm{N}(0,15) \\ \sigma_L \sim \mathrm{N}(0,15) \end{split} \end{equation} \tag{4.10}\]
As a reminder, we use the notation \(L_{[\bullet]}\) to indicate that this prior distribution applies individually to every member of the set. In our case, each of our 15 listener effects is drawn from the same normal distribution, one at a time (i.e. \(L_{[1]} \sim \mathrm{N}(0,\sigma_L), \dots , L_{[15]} \sim \mathrm{N}(0,\sigma_L)\)). In plain English, the model above says:
We expect height judgments to be normally distributed around the expected value for any given trial, \(\mu_{[i]}\), with some unknown standard deviation \(\sigma\). The expected value for a trial is equal to a fixed overall average (Intercept) and some value associated with the individual listener who made a perceptual judgment on the trial (\(L_{[\mathsf{L}_{[i]}]}\)). The listener coefficients (\(L_{[\bullet]}\)) were modeled as coming from a normal distribution with a mean of zero and a standard deviation (\(\sigma_L\)) that was estimated from the data.
There is a very important difference in how the ‘unilevel’ (complete pooling) model from Chapter 3, and this model partition the variance in apparent talker heights. The initial complete pooling model broke down the total variation in the data (\(\sigma_{total}\)) like this:
\[ \begin{equation} \sigma^2_{total} = \sigma^2 \end{equation} \tag{4.11}\]
In other words, all variation was error. We don’t know why values vary from the mean, so all variation is just random noise. Our multilevel model views the variation in our data like this:
\[ \begin{equation} \sigma^2_{total} = \sigma^2_{L} + \sigma^2 \end{equation} \tag{4.12}\]
The total variance is equal to the within-listener variance (\(\sigma^2\)) plus the between-listener variance (\(\sigma_L^2\)). From the perspective of this model, the variation within a speaker’s individual boxplot is (data-level) error. The differences from listener to listener represent random between-listener variation in apparent height judgments (i.e. listener-level error). To some extent, investigating the effects for the different listeners in our sample involves asking how much the listener averages tended to differ from the overall response. So, a crucial part of investigating the overall effect of different listeners on our results is understanding the magnitude of \(\sigma_L\).
4.5.3 Fitting the model
We can use brms to fit a model that more closely resembles our model specification in Equation 4.10. To do this we need to use the formula given above, and we also need to specify priors for three estimated parameters: \(\mathrm{Intercept}\), \(\sigma_L\), and \(\sigma\). We can confirm our expectations using the get_prior function as seen below. This time we use a clone of the function in the bmmb package simply because the output of the ‘real’ function sometimes doesn’t like it when you omit columns from the output (which we need to do so that it will fit on the page).
bmmb::get_prior (height ~ 1 + (1|L), data = men)[,-c(7:9)]
## prior class coef group resp dpar source
## student_t(3, 174.5, 7.1) Intercept default
## student_t(3, 0, 7.1) sd default
## student_t(3, 0, 7.1) sd L default
## student_t(3, 0, 7.1) sd Intercept L default
## student_t(3, 0, 7.1) sigma defaultWe see a new class of prior, sd. Our classes of priors are now:
Intercept: This is a unique class, only for intercepts.sd: This is for standard deviation parameters related to ‘batches’ of parameters, e.g.sd(Intercept)forL(\(\sigma_{L}\)).sigma: The data-level error term.
The information presented by get_prior does not just list the priors that must be specified, but also different ways that priors can be set, so there are some redundancies. This is not so necessary for our very simple models now but it will become very useful later so it is worth understanding now while it’s still relatively simple. Top to bottom, here is the information presented in the output above. First, you can specify a prior for the Intercept, which is its own unique class. Second, you can specify a prior for all sd parameters in your model. Third, you can specify a prior for all the sd parameters, specifically for your L group. The fourth line indicates that you can also specify a prior only for the Intercept for your L group (i.e. \(L\)) and not to any other listener-related predictors in your model (there are none for now). The fifth line lets you specify the prior for the error in the model (i.e. \(\sigma\)). Right now, we only have one sd parameter and so all this flexibility is not so useful, but later on, we might want to specify different priors for different sd parameters and so this will come in handy.
Below we fit a model and specify prior distributions for our intercept, for all variables in the sd class, and for the error term (basically the first, second, and fifth lines above).
# Fit the model yourself
model_multilevel = brms::brm (
height ~ 1 + (1|L), data = men, chains = 4, cores = 4,
warmup = 1000, iter = 3500, thin = 2,
prior = c(brms::set_prior("normal(176, 15)", class = "Intercept"),
brms::set_prior("normal(0, 15)", class = "sd"),
brms::set_prior("normal(0, 15)", class = "sigma")))# Or download it from the GitHub page:
model_multilevel = bmmb::get_model ('4_model_multilevel.RDS')4.5.4 Interpreting the model
We can inspect the model print statement:
# inspect model
bmmb::short_summary (model_multilevel)
## Formula: height ~ 1 + (1 | L)
##
## Group-Level Effects:
## ~L (Number of levels: 15)
## Estimate Est.Error l-95% CI u-95% CI
## sd(Intercept) 3.78 0.87 2.47 5.84
##
## Population-Level Effects:
## Estimate Est.Error l-95% CI u-95% CI
## Intercept 173.8 1.02 171.8 175.8
##
## Family Specific Parameters:
## Estimate Est.Error l-95% CI u-95% CI
## sigma 7.03 0.19 6.67 7.41This model contains one new chunk in its print statement, the Group-Level Effects, which tells us the standard deviation of the listener-dependent intercepts (sd(Intercept)) in the sample, \(\sigma_L\). Below, we calculate the listener mean height values, the standard deviation of these, and the amount of within-listener variation in apparent height judgments. We can see that these resemble the value of the analogous parameters in our model.
# find mean height for each listener
listener_means = aggregate (height ~ L, data = men, FUN = mean)
# find the within listener standard deviation
# This is the within-talker 'error'.
listener_sigmas = aggregate (height ~ L, data = men, FUN = sd)
# the mean of the listener means corresponds to our Intercept
mean (listener_means$height)
## [1] 173.8
# the standard deviation of the listener means corresponds
# to 'sd(Intercept)', the estimate of the standard deviation
# of listener intercepts
sd (listener_means$height)
## [1] 3.594
# the average within-listener standard deviation corresponds to 'sigma',
# the estimated error
mean (listener_sigmas$height)
## [1] 6.822We’re going to discuss the calculations we made above in terms of what they represent in the boxplot in Figure 4.3, our model coefficients in the print statement above, and in our model definition in Equation 4.10.
show plot code
################################################################################
par (mfrow = c(1,1), mar = c(4,4,0.5,0.5))
boxplot (height ~ L, data = men, main = "",xlab="Listener",
ylab="Apparent height (cm)",
col=bmmb::cols, ylim = c(150,195), outline=FALSE)
abline (h = mean(men$height), lwd=3,lty=3)
boxplot (height ~ L, data = men, main = "",col=bmmb::cols,add=TRUE, outline=FALSE) 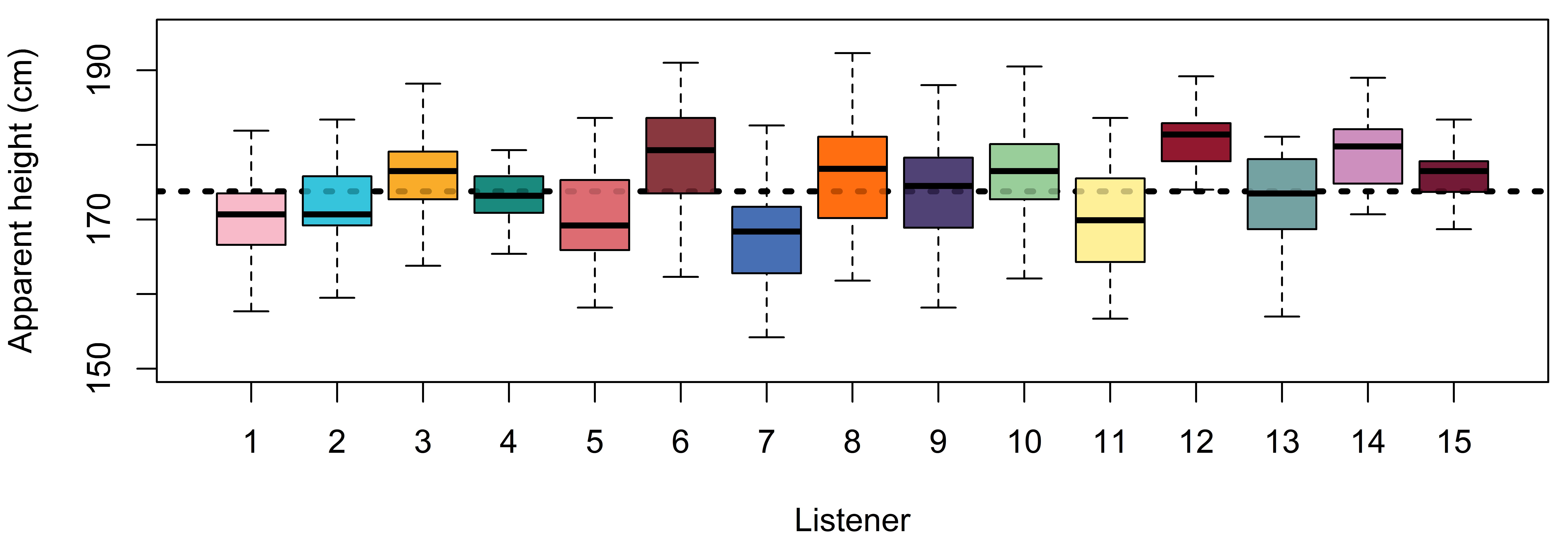
The overall mean height in our data (173.8) corresponds quite well to our model intercept of 173.8, seen in the horizontal line in the figure. The standard deviation of the listener means is 3.6 cm. This is very similar to our model estimate of 3.8 cm (sd(Intercept)). The standard deviation of the listener effects reflects the average distance from each speaker’s average to the overall average, and reflects \(\sigma_L\) in our model. Finally, the average of the within-speaker standard deviations in our data (6.8 cm) corresponds closely to our model’s error estimate (sigma = 7.0), represented by \(\sigma\) in our model. This reflects the average spread of each speaker’s data relative to their own mean, within their own little box in the figure. We can see that the information provided by brms is quite similar to what we can estimate directly using our data. However, brms does this all for us, in addition to giving us information regarding credible intervals for different parameters.
4.6 ‘Random’ effects
If you have some familiarity with statistical modeling or the analysis of experimental or repeated measures data, you may have heard of random-effects models or mixed-effects models. People often distinguish the predictors in their models between those that are ‘fixed’ and those that are ‘random’. So, a researcher might describe their model as including ‘random effects’ for so and so and ‘fixed effects’ for some other predictor. In fact, a very common way of describing our model above would be that we used a ‘mixed-effect model with random intercepts by listener’. What makes a predictor fixed or random and what is the practical effect of this in our models? It turns out that although they are commonly used, the terms ‘fixed’ and ‘random’ effects do not have a single agreed-upon definition. Gelman (2005) highlights five different definitions of the terms given in the literature:
Fixed effects are constant across individuals/groups, while random effects can vary across them.
Effects are fixed when you are interested in them specifically, but random if you are actually interested in the population.
When a sample represents the whole population, the effect is fixed. If the sample is a small part of the population, the effect is random.
If the effect is assumed to be an observation from a random variable, it is a random effect.
Fixed effects are estimated using maximum likelihood and no pooling. Random effects are estimated using partial pooling (and shrinkage).
You will note that some of these definitions would have an effect change from random to fixed based on the researcher’s conception of it. Rather than focus on effects as fixed or random in theory, we may ask: What is the difference between these sorts of effects in practice? What special treatment are we referring to when we say that a certain predictor in our model is a ‘random effect’? The answer is that when researchers discuss the inclusion of random effects in their models, what they are usually (if not always) referring to is that the effects were estimated using adaptive partial pooling. When researchers indicate that their effects were fixed effects, they are usually saying that they were fit with no, or at least minimal, pooling (i.e. using flat or weak priors). This applies to both Bayesian models, and to models fit using more ‘traditional’ approaches such as the popular lmer function in the lme4 package in R (Bates et al. 2015).
As a result, although researchers may define ‘random’ effects using any of the definitions above, in practice, terms labeled ‘random’ are usually those estimated using adaptive partial pooling. In light of this, Gelman and Hill (2006) recommend avoiding the terms ‘fixed’ and ‘random’, since these are vaguely defined and overly-broad. Although we agree with this position, the fact is that in many research areas it is still most common to refer to effects as being either fixed or random. For that reason, we will follow this convention, although we will continuously highlight the relevant underlying distinction: That so-called ‘random’ effects are simply those that are estimated using adaptive partial pooling.
In general, when you have many levels of a factor, it may be a good idea to include these as ‘random’ effects, regardless of how ‘random’ they might actually be. There is not much downside to it: You get more information from the model (e.g., information about \(\sigma_{predictor}\)), there are several modeling advantages associated with estimating large batches of coefficients with adaptive pooling (see chapter 11), and you can always fit multiple models to see what, if any differences, exist across the two approaches. Rather than considering the more ‘philosophical’ position outlined in 1–5 above, some useful things to consider when deciding between fixed and random effects are: Do you believe that treating a predictor as a ‘random’ effect offers a modeling advantage? Does it better reflect the process/relationships you’re trying to understand? Does it provide you with information you find useful? Is it realistic to fit this kind of model given the amount of data you have, and the nature of the relationships in the data? Right now the last point is not something we’ve talked about very much, but it is something we will need to worry about as our models become more complicated.
4.6.1 Inspecting the random effects
You may have noted that the estimates of our coefficients fit with adaptive pooling (\(L\)) do not appear in the model print statement. Instead, only the standard deviation of the effects is presented (\(\sigma_L\), or sd(Intercept)). The reason for this is that there may sometimes be a very large number of random effects coefficients, which would result in models that are difficult to read if they were all shown. A second reason for this is that researchers are often not directly interested in the individual values of their random effects, but rather they are usually focused on the overall effects common to all listeners such as the model intercept. For example, to this point we have been talking about “what is the average apparent height of male speakers” and never “what did listener 04 think the average height of speaker 101 is?”. However, there are situations in which we may be interested in investigating the random effects, and the brms package has several functions to facilitate this process.
We will present two ways to consider the random listener effects using the ranef function, which gets information about your random effects from your brms model. In the first of these, you set summary=FALSE in the call to ranef as shown below. This function returns a list of three-dimensional matrices, where each list element is named after the grouping factor it represents. Below, we collect the random effects and take the element named L, corresponding to a matrix representing our listener factor.
# get random effects, without summarizing
random_effects = brms::ranef (model_multilevel, summary = FALSE)
# get only the listener random effects
listener_effects = random_effects$LThe matrix representing each factor has three dimensions, with individual posterior samples varying across the first dimension (rows), factor levels varying along the second dimension (columns), and parameters varying across the third dimension. Basically, listener_effects is a set of two-dimensional matrices stuck together, one for each random parameter. Of course, this time we only have a single random effect for listeners, so there is only one element along the third dimension.
str (listener_effects)
## num [1:5000, 1:15, 1] -1.76 -2.74 -1.39 -6.66 -2.71 ...
## - attr(*, "nchains")= int 4
## - attr(*, "dimnames")=List of 3
## ..$ : NULL
## ..$ : chr [1:15] "1" "2" "3" "4" ...
## ..$ : chr "Intercept"You can select the matrix corresponding to the intercept by putting the name of the parameter in the third dimension of the matrix, as seen below. This reduces our three-dimensional matrix to a two-dimensional one representing the random listener intercepts in our model.
# take only the intercept along the third dimension
listener_intercepts = listener_effects[,,"Intercept"]The above process results in a two-dimensional matrix representing the individual samples of our listener effects. We can see below that this matrix has 5,000 rows, corresponding to the individual samples, and 15 columns, representing the individual listeners. We use the head function to see the first six samples of the listener effects. To be totally clear, each column represents the samples of \(P(L_{[\bullet]} | y)\) for different speakers. So, the first column represents the individual posterior samples of the listener effect for listener 1 (\(P(L_{[1]} | y)\)), and the second column represents the individual posterior samples of the listener effect for listener 2 (\(P(L_{[2]} | y)\)),
# Our matrix of posterior samples showing 5000 samples
# for each of 15 listener effects
dim (listener_intercepts)
## [1] 5000 15
# the first six samples of the listener effect for 6 listeners
head ( round (listener_intercepts[,1:6], 2))
## 1 2 3 4 5 6
## [1,] -1.76 -2.12 3.24 1.29 -2.95 3.13
## [2,] -2.74 -1.65 1.84 0.00 -2.93 5.85
## [3,] -1.39 -2.99 2.47 -0.02 -5.25 2.98
## [4,] -6.66 -5.17 -2.59 -1.72 -6.98 1.72
## [5,] -2.71 -2.13 3.70 -0.83 -3.61 3.89
## [6,] -3.20 -1.86 1.96 -1.74 -3.67 5.17If we were to find the average of the samples within each column, the result would be the posterior mean estimate for each of our individual listener effects.
colMeans (listener_intercepts)[1:10]
## 1 2 3 4 5 6 7 8 9 10
## -3.61 -2.65 1.49 -0.69 -3.80 3.81 -5.52 1.91 0.25 1.98We can repeat the process above but without specifying summary=FALSE. When you do this, rather than the individual samples you get summaries of the posterior distribution of each parameter. We will still get a list of three-dimensional matrices. However, this time factor levels vary across the first dimension (rows), information about each factor level varies across columns, and parameters vary across the third dimension. We again take the list element corresponding to listeners (L) and take only the third dimension corresponding to our Intercept.
# get random effects, *with* summarizing (by default)
random_effects = brms::ranef (model_multilevel)
# get only the listener random effects
listener_effects = random_effects$L
# along the third (coefficient) dimension, take only the intercept dimension
listener_intercepts = listener_effects[,,"Intercept"]This time, the process results in a two-dimensional matrix representing a summary of the posterior, as seen below. The leftmost column of the output below represents the posterior mean effect estimated for each listener, the second column represents the standard deviation of posterior samples, and the third and fourth columns provide he 2.5% and 97.5% credible intervals for each parameter, respectively. We can see that the information in the first column below exactly matches the posterior means we calculated for each listener effect above. This is because they are the same thing. In the un-summarized representation above we were seeing the individual samples varying across columns, and we ‘summarized’ them by averaging across the columns. In the summarized representation we are seeing the same samples, summarized in each row.
listener_intercepts[1:6,]
## Estimate Est.Error Q2.5 Q97.5
## 1 -3.6106 1.404 -6.388 -0.88497
## 2 -2.6477 1.387 -5.419 0.07352
## 3 1.4902 1.387 -1.266 4.27533
## 4 -0.6855 1.386 -3.393 2.04369
## 5 -3.7971 1.382 -6.494 -1.11026
## 6 3.8118 1.399 1.050 6.60174Notice that the listener averages vary around 0, and some are even negative. That is because the listener effects (and all random effects) are represented as deviations from the intercept, as noted above. We can verify this by calculating the average reported height for each listener and centering these values. We can then compare these centered means to our model random effects and see that they are very similar.
# find listener means
listener_means = tapply (men$height, men$L, mean)
# center them
listener_means_centered = listener_means - mean(listener_means)
# compare centered means (top row) to model random effects (bottom row)
round (rbind (listener_means_centered,
listener_random_effects = listener_intercepts[,1]))
## 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15
## listener_means_centered -4 -3 2 -1 -4 4 -6 2 0 2 -4 7 -1 3 2
## listener_random_effects -4 -3 1 -1 -4 4 -6 2 0 2 -3 6 -1 3 1We can use a simple plotting function included in the bmmb package (brmplot) to inspect the posterior distributions of our model coefficients. The function takes in the summary matrices made by brms and plots a point for each parameter mean/median, and lines indicating the credible intervals calculated by brm (usually 95% credible intervals). These matrices all have a standard form where parameters vary across rows, the first column is the posterior mean, the second is the posterior standard deviation (the estimated error), and the third and fourth columns are the 2.5% and 97.5% credible intervals. If you have a matrix like this you can easily plot it with brmplot as shown below.
bmmb::brmplot(listener_intercepts)We can compare the estimates of our by-listener effects to the distribution of centered height responses arranged by subject. In Figure 4.4 we plot centered listener responses using boxplots and then use brmplot to overlay the colored points and lines, representing the listener ‘random effects’, on top. We can see that the estimated listener parameters and the raw data show a close correspondence.
show plot code
################################################################################
par (mfrow = c(1,1), mar = c(4,4,1,1))
height_centered = (men$height-mean(men$height))
boxplot (height_centered ~ L, data=men, col = 0, ylim = c(-23,21), xlab="Listener",
ylab="Centered apparent height (cm)", outline = FALSE)
abline (h=0)
bmmb::brmplot(listener_intercepts, col = bmmb::cols, ylim = c(-30,20),
add = TRUE, outline = FALSE)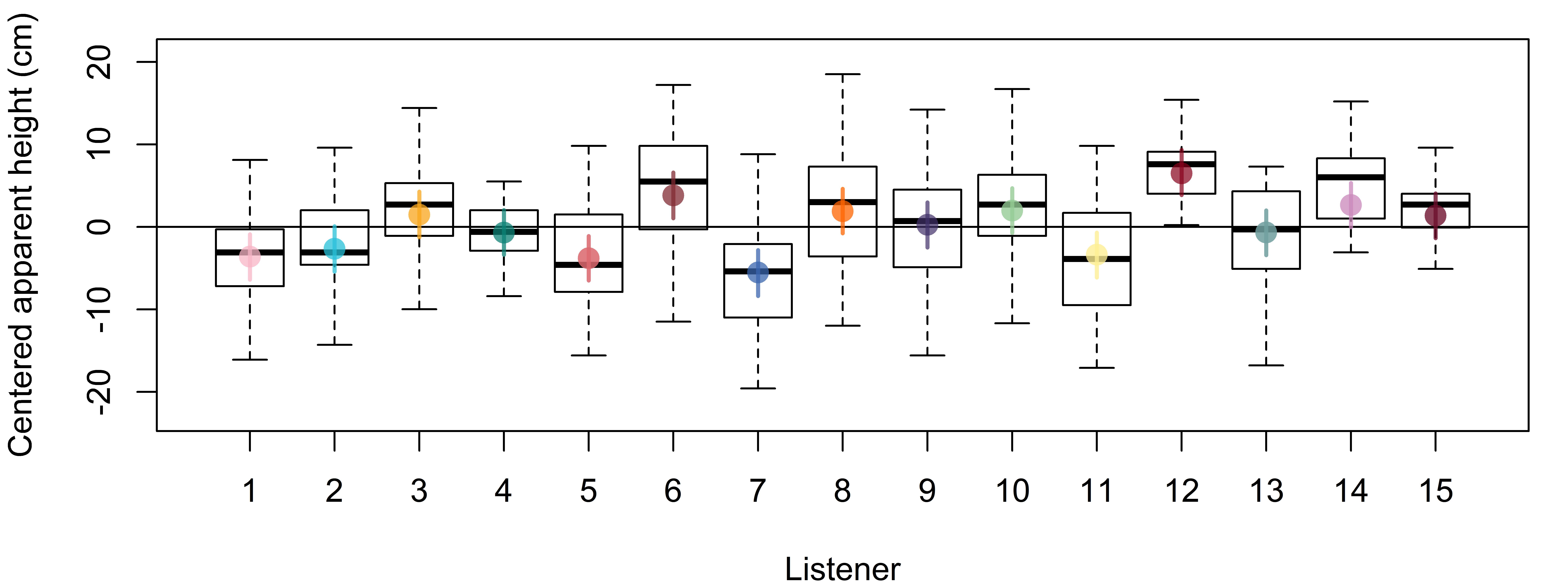
4.7 Simulating data using our model parameters
Regression models break up variation in the dependent variables into component parts. One way to think about what all the numbers in our model mean is to simulate the dependent variable by building it from the individual component parts indicated by the model. Our model (model_multilevel) has just four components we need to include, one for every estimated parameter: The intercept, the listener-dependent intercepts (\(L\)), the between-listener variation (\(\sigma_L\)), and the within-listener variation, the error (\(\sigma\)).
We do this in the code below. First we set the intercept to 174 cm. Then, we sample 15 random listener effects from a normal distribution, representing simulated listeners. The listener population has a mean of 0 and a standard deviation (\(\sigma_L\)) equal to 3.8 as indicated by our model. These effects are stored in a vector called L_, and they represent the values of \(L\) for our different listeners. After this, we create an index vector L that tells us which value of L_ to apply for any given trial. This vector contains values from 1 to 15, representing each listener, repeated 45 times each in order to simulate 45 observations from each listener. Finally, we draw our error (i.e. \(\varepsilon \sim \mathrm{N}(0,\sigma)\)) based on the magnitude of sigma in our model. Note that the error is just \(45 \cdot 15\) random draws from this population. Since the model contains the same value of \(\sigma\) for all trials, there is no distinction between one listener and another when it comes to the magnitude of the error.
# skip this line if you want a new simulated data set.
set.seed(1)
# this is the value of our intercept
Intercept = 174
# this is a vector of 15 listener effects
L_ = rnorm (15, 0, 3.8 )
# this is a vector indicating which listener produced which utterance
L = rep (1:15, each = 45)
# this vector contains the error
error = rnorm (45 * 15, 0, 7)After creating our components, we add the Intercept, listener effects, and random error to make our fake ‘replicated’ data. Since this data has the same statistical properties as our real data, it should look a lot like it.
# the sum of an intercept, listener effects and random error
height_rep = Intercept + L_[L] + errorIn Figure 4.5 we compare the results of our simulation to our real data. The are reasonably similar, which tells us that our model is a good reflection of the data.
show plot code
################################################################################
par (mfrow = c(1,2), mar = c(0.5,0.5,1,0.5), oma = c(0,3.5,0,0))
boxplot (height_rep ~ L, ylim = c(140,200), xaxt='n',xlab='',
col=bmmb::cols)
abline (h = 173.8, lwd=3,lty=3)
boxplot (height ~ L, data = men, ylim = c(140,200), xaxt='n',xlab='',
col=bmmb::cols,yaxt="n")
abline (h = 173.8, lwd=3,lty=3)
mtext (text = "Apparent height (cm)", side=2, outer=TRUE, line = 2.5)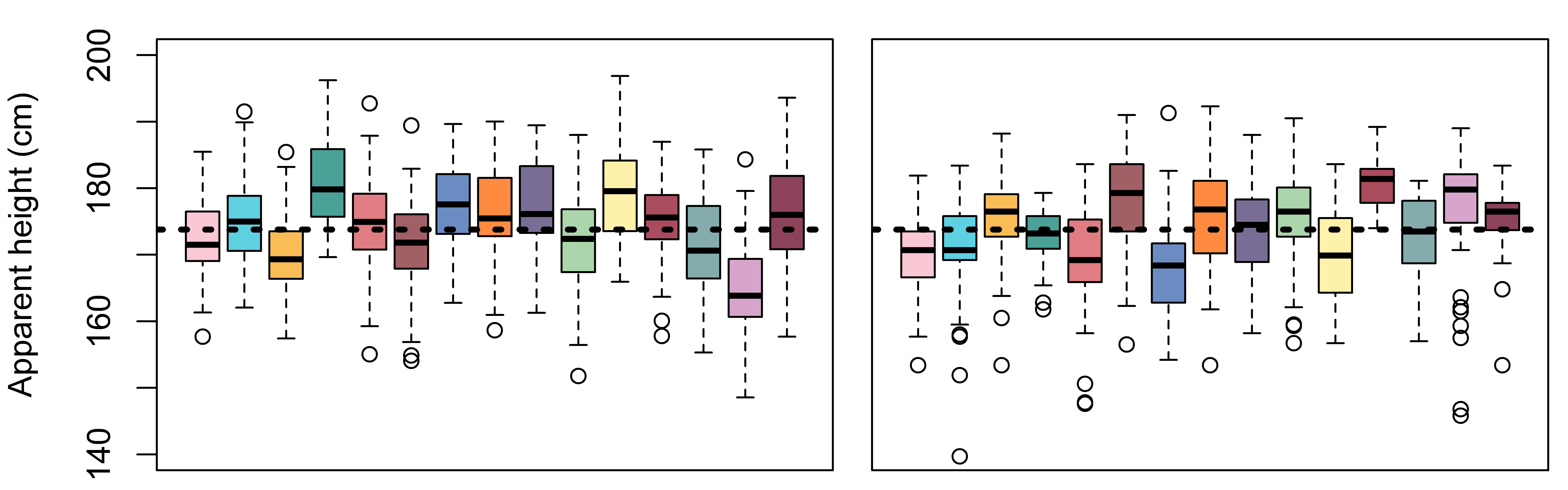
Below we make two data sets that are ‘incomplete’ with respect to the variation in our data. The first contains the intercept and noise only, the second contains the intercept and listener effects only.
# this fake data is missing between listener variation
height_rep_1 = Intercept + error
# this fake data is missing within listener variation
height_rep_2 = Intercept + L_[L]In Figure 4.6 we compare these ‘incomplete’ data sets to the full simulated data. The top row contains only error (\(\sigma\)). As a result, height judgments vary around the intercept, but there is no listener-to-listener variation. This is what that data would look like if there is no variation in means across listeners, as is assumed by the ‘unilevel’ (complete pooling) model in Figure 4.2. However, notice that each little box is not centered at zero. Although we have not added any variation in listener means, a small amount of this appears to be present in our data anyways. This is because although the error distribution has a mean of zero, each of our listener boxes contains only a small sample of errors. This tiny sample of errors is extremely unlikely to add up to exactly zero. Whatever the errors do add up to, this value will contribute some unknown amount to each listener’s sample mean estimate. This makes estimation of the real listener effects impossible in practice.
show plot code
################################################################################
par (mfrow = c(3,1), mar = c(.1,3,.1,1), oma = c(1,2,1,0))
boxplot (height_rep_1 ~ L, ylim = c(140,198),xaxt='n',
col=bmmb::cols,ylab="",yaxt = 'n')
text (1, 145, label = "Intercept + error", cex = 1.5, pos=4)
abline (h=174.8,lty=2)
axis (side=2,at=seq(150,190,10))
boxplot (height_rep_2 ~ L, ylim = c(140,198),xaxt='n',
col=bmmb::cols,ylab="",yaxt = 'n')
abline (h=174.8,lty=2)
text (1, 145, label = "Intercept + L", cex = 1.5, pos=4)
axis (side=2,at=seq(150,190,10))
boxplot (height_rep ~ L, ylim = c(140,198),xaxt='n',
col=bmmb::cols,ylab="",yaxt = 'n')
abline (h=174.8,lty=2)
text (1, 145, label = "Intercept + L + error", cex = 1.5, pos=4)
mtext (side=2,text="Apparent height (cm)", outer = TRUE, line=0)
axis (side=2,at=seq(150,190,10))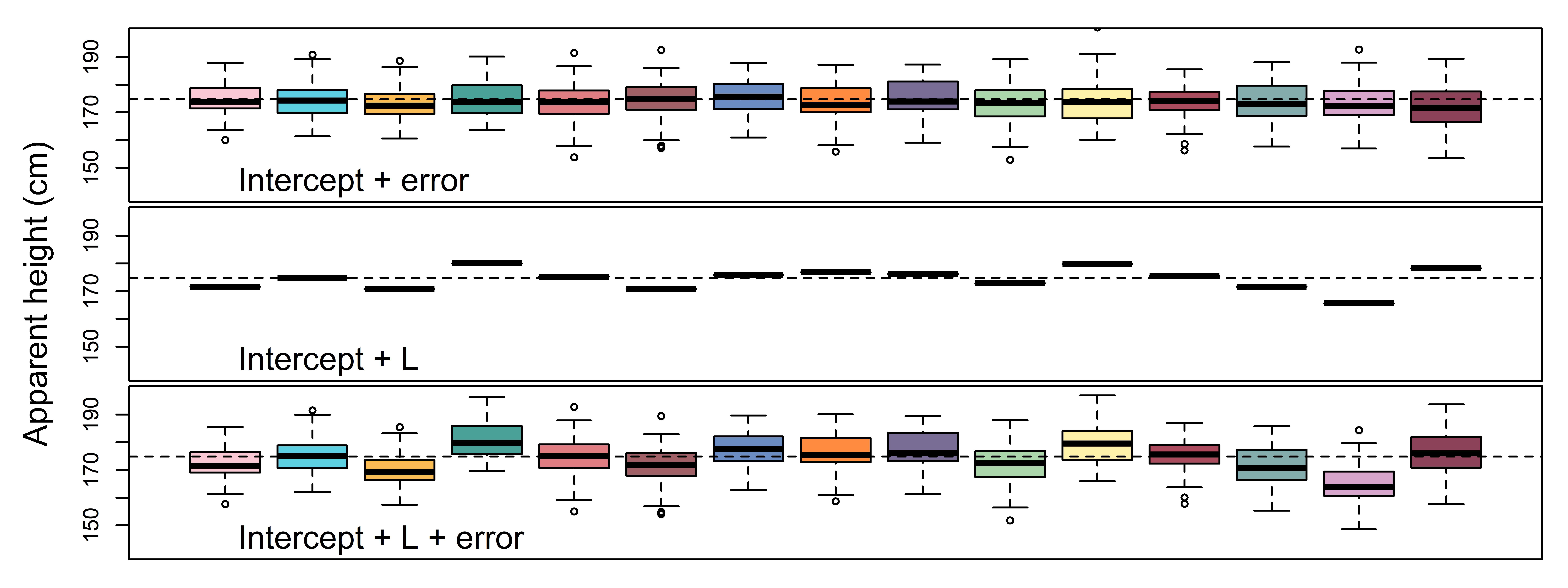
In the middle plot, the figure shows only between-listener variation (\(\sigma_{L}\)) but no within-listener variation (i.e. no \(\sigma\)). Now listeners vary from the intercept, and between each other, but they do not vary within themselves. We can see the effect of this, the boxes have no ‘width’, they are lines. That is because in this situation listeners would always respond at their mean, showing no internal variation in their responses. The final plot is the combination of the variation in the top two plots and shows the final simulated data: The sum of the intercept, the within-listener variation, and the between-listener variation.
As seen in the top row of Figure 4.6, the model without listener variation is unlikely to generate the substantially-different listener boxplots we see in our data through noise alone. However, they do show a tiny bit of variation in the listener means due to error alone (as noted above). So, random error can give the impression of listener variation even when none exists. We can investigate how likely our error is to generate listener means as diverse as those we have observed, by accident. To do this, we run one more simulation, identical in all respects except for two things: (1) We set the between-listener variance to zero, and (2) we set the noise variance to 7.8 (as in our complete pooling model). Below we generate 10,000 simulated data sets and record the standard deviation of the listener means for each one. These values will be stored in a vector called sigma_L_rep since we are simulating values of \(\sigma_L\) for our complete pooling model.
set.seed(1)
# do 10,000 replications
reps = 10000
# hold the replicated values of sigma_L
sigma_L_rep = rep(0,reps)
for ( i in 1:reps){
Intercept = 173.8 # set the intercept
L_L = rnorm (15, 0, 0) # zero between-listener variance
L = rep (1:15, each = 45) # 45 responses from each of 15 listeners
epsilon = rnorm (45 * 15, 0, 7.78) # generate random noise
height_rep = Intercept + L_L[L] + epsilon # add up to components
L_rep_means = tapply(height_rep, L, mean) # get replicated listener means
sigma_L_rep[i] = sd (L_rep_means) # find sigma of listener effects
}Below, we see that even in 10,000 simulations we do not see a standard deviation greater than 2.1, and that 75% are smaller than 1.3.
quantile(sigma_L_rep)
## 0% 25% 50% 75% 100%
## 0.4429 0.9862 1.1337 1.2810 2.0813The standard deviation of the listener means in our data was 3.6 and our model estimate of \(\sigma_L\) was 3.8. This reinforces the idea that between-listener variation (\(\sigma_L\)) is a ‘real’ aspect of our data, which is extremely unlikely to arise incidentally due to random variation, and that our models will benefit from taking it into account.
4.8 Adding a second random effect
Our model is looking better but is still not ‘right’. This is because, in addition to repeatedly sampling from only 15 listeners, our model also features repeated sampling from only 45 different speakers. In Figure 4.7 we see that we can recreate Figure 4.1, this time illustrating systematic variation between speakers in addition to between listeners. So, we are interested in the average apparent height of adult males overall but have only sampled 45 of them, and we need to build this information into our model.
show plot code
################################################################################
library (bmmb)
data (exp_data)
men = exp_data[exp_data$C_v=='m',]
# create vector of height judgments
mens_height = men$height
#par (mar = c(.1,.1,.5,.1), mfrow = c(1,1), oma = c(4.2,.20,0,.2))
#layout (m=matrix(c(1,2,3,4),2,2), heights = c(.25,.75))
par (mar = c(4.5,.5,.5,.5), mfrow = c(1,3))
#plot (0,type='n',xlab='',ylab='',xaxt='n',yaxt='n',bty='n')
boxplot (men$height,col = 'grey', ylim = c(140,195),
width = 2, ylab="", horizontal = TRUE)
mtext (side=2,outer=TRUE, text="Apparent height (cm)", cex=1, line=2.9,adj=.55)
#boxplot (tapply(men$height,men$L,mean), xlab = "",xaxt='n',
# col = cols, ylim = c(140,195), width = 2,cex.axis=1.3,
# horizontal = TRUE)
#abline (h=)
#text (1,190,expression(sigma["[within]"]), cex=2)
boxplot (height ~ L, data = men, xlab = "", ylab="",xaxt='s',
col = cols[-1], ylim = c(140,195), cex.lab=1.1,cex.axis=1.1,
horizontal = TRUE, yaxt='n')
grid()
abline (h = mean(mens_height), lwd=2, lty=3)
boxplot (height ~ L, data = men, col = cols[-c(1,8)],add=TRUE,yaxt='n',xaxt='n')
boxplot (height ~ S, data = men, xlab = "", ylab="",xaxt='s',
col = cols[-1], ylim = c(140,195), cex.lab=1.1,cex.axis=1.1,
horizontal = TRUE, yaxt='n')
grid()
abline (h = mean(mens_height), lwd=2, lty=3)
boxplot (height ~ S, data = men, col = cols[-c(1,8)],add=TRUE,yaxt='n',xaxt='n')
mtext (side=1,outer=TRUE, "Apparent height (cm)",line=-1.3)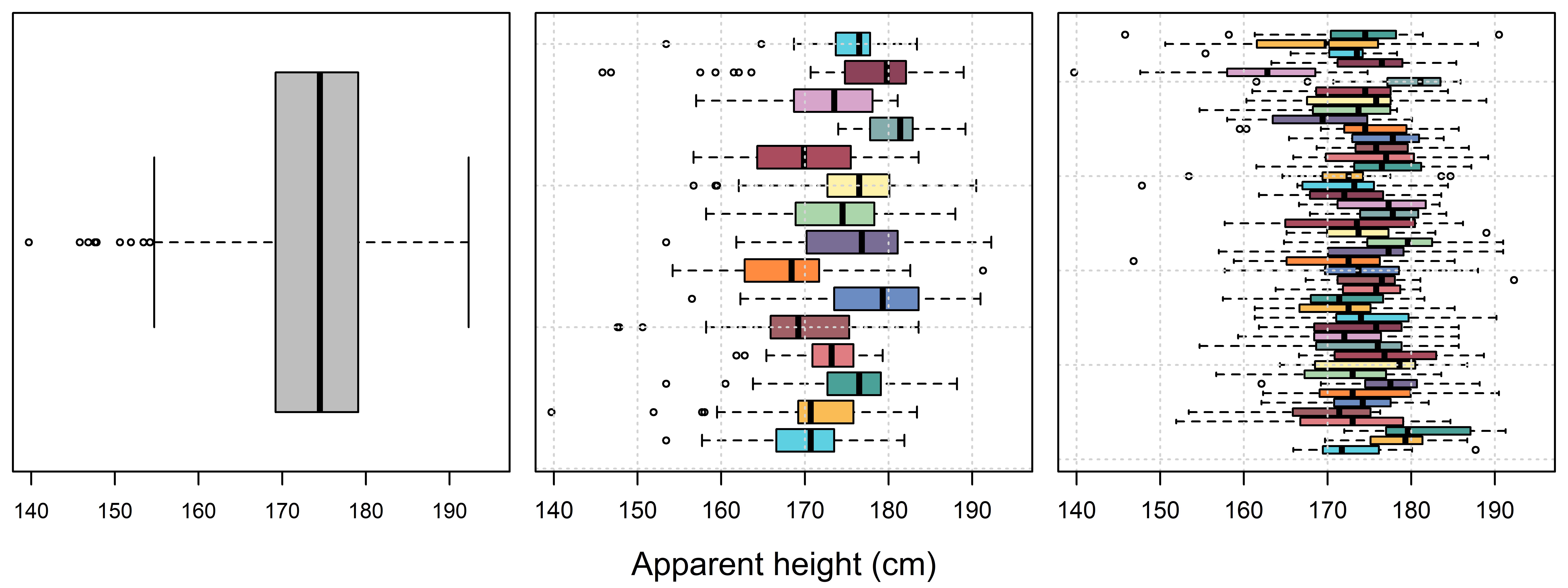
In order to investigate this, our model will now include the following additional variable from our exp_data data frame:
S: An integer from 1 to 139 indicating which speaker produced the trial stimulus.
4.8.1 Updating the model description
In order to model the effects of the different speakers in our data, our formula must be updated to include our S (speaker) predictor in parentheses, like this:
height ~ 1 + ( 1 | L) + ( 1 | S)
The new term in parentheses indicates a second predictor whose levels we want to estimate using adaptive pooling. Again, we include only a one on the left-hand side of the pipe indicating that we are only estimating speaker-specific intercepts. So, this formula says ‘predict height using only an overall intercept, but also estimate a different intercept for each level of listener and speaker’. The regression model corresponding to the formula above now looks like this:
\[ \begin{equation} \begin{split} height_{[i]} \sim \mathrm{N}(\mu_{[i]},\sigma) \\ \mu_{[i]} = \mathrm{Intercept} + L_{[\mathsf{L}_{[i]}]} + S_{[\mathsf{S}_{[i]}]} \\ \\ \mathrm{Priors:} \\ L_{[\bullet]} \sim \mathrm{N}(0,\sigma_L) \\ S_{[\bullet]} \sim \mathrm{N}(0,\sigma_S) \\ \\ \mathrm{Intercept} \sim \mathrm{N}(176,15) \\ \sigma \sim \mathrm{N}(0,15) \\ \sigma_L \sim \mathrm{N}(0,15) \\ \sigma_S \sim \mathrm{N}(0,15) \end{split} \end{equation} \tag{4.13}\]
Just as with the listener coefficients (\(L\)), the speaker coefficients (\(S\)) represent deviations in values from the intercept and so the speaker average is set to zero. In plain English, the model description above says:
We expect height judgments to be normally distributed around the expected value for any given trial, \(\mu_{[i]}\), with some unknown standard deviation \(\sigma\). The expected value for a trial is equal to the sum of a fixed overall average (Intercept), some value associated with the individual listener (\(L_{[\mathsf{L}_{[i]}]}\)) who judged the trial, and some value associated with the individual speaker (\(S_{[\mathsf{S}_{[i]}]}\)) who produced the trial. The listener and speaker coefficients (\(L\) and \(S\)) were both modeled as coming from normal distributions whose standard deviations, \(\sigma_L\) and \(\sigma_S\), were estimated from the data. Prior distributions for \(\sigma\), \(\sigma_L\), \(\sigma_S\), and the intercept were all specified a priori.
The model we fit in Chapter 3 only included variation due to error. The first model we fit in this chapter divided variation into random between-listener variation and random within-listener error. Now, our model breaks up the total variance in our data into three components: Between-listener variation, between-speaker variation, and random error, as seen below.
\[ \begin{equation} \sigma^2_{total} = \sigma^2_{L} + \sigma^2_{S} + \sigma^2 \end{equation} \tag{4.14}\]
If the error was the within-listener variation before, what does it represent now? Our error is now the difference between the value we expect, given the speaker and listener, and the value we observe. So, it is the variation adjusting for the speaker and listener. In other words, if you know who the speaker is and who the listener is, and you adjust for this, how much random variability do you still have left over in your model? The answer is \(\sigma\).
4.8.2 Fitting and interpreting the model
Below, we fit the new model using exactly the same code as for model_multilevel, save for the modification to the model formula. We don’t need to specify a hyperprior for \(\sigma_S\) because this falls under the sd category (see Section 4.5.3). As long as we can use the same hyperprior for each factor-specific standard deviation, i.e. \(\sigma_{F}\) for factor \(F\), we don’t need to specify a prior for each parameter. We think a prior expectation of 15 cm is reasonable for the speaker standard deviation given that the standard deviation of adult male heights in the US is around 7.5 cm.
# Fit the model yourself
model_multilevel_L_S = brms::brm (
height ~ 1 + (1|L) + (1|S), data = men, chains = 4, cores = 4,
warmup = 1000, iter = 3500, thin = 2,
prior = c(brms::set_prior("normal(176, 15)", class = "Intercept"),
brms::set_prior("normal(0, 15)", class = "sd"),
brms::set_prior("normal(0, 15)", class = "sigma")))# Or download it from the GitHub page:
model_multilevel_L_S = bmmb::get_model ('4_model_multilevel_L_S.RDS')After fitting the model we inspect the output, some of which we include below:
bmmb::short_summary (model_multilevel_L_S)## ~S (Number of levels: 45)
## Estimate Est.Error l-95% CI u-95% CI
## sd(Intercept) 2.83 0.42 2.1 3.72The model print statement should look familiar except it contains one new chunk representing information about the speaker predictor in our model (shown above). This chunk reminds us that our S predictor had 45 levels, and provides us information about the estimate of the \(\sigma_S\) parameter (sd(Intercept)), which is 2.83 according to our model.
4.9 Investigating ‘shrinkage’
We’ve mentioned that multilevel models can result in shrinkage, that is, estimates that are smaller in magnitude (i.e. closer to the mean) than their no pooling counterparts. In the code below we extract the posterior mean estimates of our listener and speaker random effects (i.e. \(L\) and \(S\)). These estimates have been ‘shrunken’ by combining their likelihoods with the prior. We also calculate the no pooling estimates for the listener and speaker averages, and center these so that they will resemble their corresponding effects.
# these are the Bayesian 'shrinkage' estimates of
# the listener effects
L_shrunk = brms::ranef(model_multilevel_L_S)$L[,1,'Intercept']
# and the speaker effects
S_shrunk = brms::ranef(model_multilevel_L_S)$S[,1,'Intercept']
# these are the no pooling estimates
L_nopooling = tapply (men$height, men$L, mean)
S_nopooling = tapply (men$height, men$S, mean)
# and now they are centered
L_nopooling = L_nopooling - mean (L_nopooling)
S_nopooling = S_nopooling - mean (S_nopooling)Figure 4.8 shows the result of shrinkage for the listener and speaker effects: Individual effects are ‘pulled’ toward the overall mean at zero. We can see that in all cases, the effects estimated with adaptive pooling lie closer to the mean than those estimated with no pooling. This is particularly evident for the speaker effects, and in particular, those of speaker 87, highlighted in that figure.
show plot code
################################################################################
par (mfrow = c(1,2), mar = c(2,.51,.5,.51))
layout (mat = matrix(c(1,3,2,3), 2,2))
plot (L_nopooling,rep(0,15), ylim = c(0,4.3), cex=1.5,pch=1,col=rev(bmmb::cols), xlim = c(-13,13),
yaxt = 'n',xlab = 'Listener Effects',lwd=3)
points (L_shrunk,rep(1,15), cex=1.5,pch=16,col=rev(bmmb::cols),lwd=2)
arrows (L_nopooling,0.1,L_shrunk,0.9, length=0.1)
abline (v=0, lty=3,col='grey')
x = seq(-12,12,0.01)
den = dnorm (x, 0, 3.79)
den = den / max (den)
den2 = dnorm (x, 0, sd(S_nopooling))
den2 = den2 / max (den2)
text (0,2.5,label = expression(paste(sigma["L"]," = 3.79")), cex = 1.25)
text (0,3,label = expression(paste("P(L)")), cex = 1.25)
lines (x, (den*2)+2, lwd = 3, col = 4)
#lines (x, (den2*2)+2, lwd = 2, col = 4,lty=3)
text (-13.5,0,"No pooling", pos = 4)
text (-13.5,1,"Shrunken", pos = 4)
text (5,4,"Listener Effects", pos = 4)
plot (S_nopooling,rep(0,45), ylim = c(0,4.3), cex=1.5,pch=1,col=rev(bmmb::cols), xlim = c(-13,13),
yaxt = 'n', ylab = '',xlab = 'Speaker Effects',lwd=3)
points (S_shrunk,rep(1,45), cex=1.5,pch=16,col=rev(bmmb::cols),lwd=2)
arrows (S_nopooling,0.1,S_shrunk,0.9, length=0.1)
abline (v=0, lty=3,col='grey')
x = seq(-12,12,0.01)
den = dnorm (x, 0, 2.84)
den = den / max (den)
den2 = dnorm (x, 0, sd(L_nopooling))
den2 = den2 / max (den2)
text (0,2.5,label = expression(paste(sigma["S"]," = 2.84")), cex = 1.25)
text (0,3,label = expression(paste("P(S)")), cex = 1.25)
lines (x, (den*2)+2, lwd = 3, col = 4)
#lines (x, (den2*2)+2, lwd = 2, col = 4,lty=3)
text (5,4,"Speaker Effects", pos = 4)
text (-12, 0.5, "S=87")
plot (S_nopooling[41],0.05, ylim = c(0,1.5), cex=2.5,pch=1,
col=bmmb::cols[5], xlim = c(-17,9), yaxt = 'n', ylab = '',
xlab = 'Speaker Effects',lwd=3, yaxs = 'i')
points (S_shrunk[41],0.05, cex=2.5,pch=16,col=bmmb::cols[5],lwd=2)
arrows (S_nopooling[41],0.05,S_shrunk[41]-.5,0.05, length=0.1)
x = seq(-20,15,0.01)
den = dnorm (x, 0, 2.84)
den = den / max (den)
tmp = exp_data$height[exp_data$S==87] - 173.8
den2 = dnorm (x, mean (tmp), 6.47/sqrt(14))
den2 = den2 / max (den2)
den3 = den2 * den
den3 = den3 / max (den3)
den4 = dnorm (x, -8.560 , 1.648)
den4 = den4 / max (den4)
use2 = (x > -17) & (x < -5)
lines (x[use2], (den2)[use2], lwd = 3, col = bmmb::cols[5], lty = 2)
#lines (x[use2], (den4)[use2], lwd = 3, col = bmmb::cols[6], lty = 2)
use3 = (x > -13) & (x < -2)
lines (x[use3], (den3[use3]), lwd = 3, col = bmmb::cols[5])
use = (x > -10) & (x < 10)
lines (x[use], (den[use]), lwd = 3, col = 4)
points (S_nopooling[41],0.05, cex=2.5,pch=1,col=bmmb::cols[5])
points (S_shrunk[41],0.05, cex=2.5,pch=16,col=bmmb::cols[5],lwd=2)
text (S_nopooling[41],1.25,label = expression(paste("P(y|",S["[87]"],")")), cex = 1.25)
text (S_shrunk[41],1.25,label = expression(paste("P(",S["[87]"],"|y)")), cex = 1.25)
text (0,1.25,label = expression(paste("P(",S,")")), cex = 1.25)
text (S_nopooling[41],.5,label = "Likelihood", cex = 1.25)
text (S_shrunk[41],.5,label = "Posterior", cex = 1.25)
text (0,.5,label = "Prior", cex = 1.25)
abline (v=0,lty=3,col="grey")
In the bottom row of Figure 4.8, we focus on the process by which the effect for speaker 87 gets shrunken from a no pooling value of −11.6 to a shrunken (adaptive pooling) value of −8.6. This example is simply meant to illustrate how this happens at a conceptual level, the actual calculations and processes involved are more complicated than this. Figure 4.8 is very similar to Figure 3.1, which presented the blending of priors and likelihoods to find the posterior. The shrinkage of random effects arises from the same behavior: We combine the likelihood of a parameter in isolation with prior information about other parameters like it to arrive at a posterior estimate that blends the two.
Our ‘no pooling’ estimate is an estimate based solely on the individual likelihood of the parameter. The likelihood function we present in the figure is the likelihood of the speaker effect for speaker 87 given their judged height on 15 observations. To calculate this, we assume that \(\sigma=6.47\) as in our last model, and use centered height judgments. The prior density we present in the figure is based on our model parameters, it has a mean of 0 and a standard deviation of 2.83. To find the posterior distribution presented in the bottom row of Figure 4.8, we multiplied the curves representing the likelihood and the prior for each point along the x-axis. So, even in this simple demonstration we can see how effect estimates get pulled, or shrunken, away from their no pooling values by their priors.
In Figure 4.9 we plot the no pooling estimates (x-axis) against the difference between the no pooling and shrunken estimates (y-axis). The difference between the no pooling and shrunken estimate is basically the magnitude of the horizontal arrow shown in the bottom row of Figure 4.8, for each pair of shrunken and no pooling parameters. It is the amount of tilt in the arrows in the top row of the same figure; in the absence of shrinkage they would all be vertical. We can see that the amount of shrinkage, the difference between the no pooling and shrunken estimates, varies as a function of the magnitude of the estimate. More deviant estimates are shrunken more, and less deviant estimates are shrunken less.
show plot code
################################################################################
par (mfrow = c(1,2), mar = c(.25,4,.25,1), oma=c(4,.1,.1,.1))
#
# plot (L_nopooling, L_shrunk, xlim = c(-10,10), ylim = c(-10,10),
# pch=4, lwd=3, col=bmmb::cols,cex=2.25,xaxt="n",xlab="")
# grid()
# abline (0,1,col=2,lwd=2)
# abline(h=0,v=0,lty=3,lwd=2)
# points (L_nopooling, L_shrunk, pch=4,lwd=3, col=bmmb::cols,cex=2.25)
#
# plot (S_nopooling, S_shrunk, xlim = c(-12,12), ylim = c(-12,12),
# pch=4, col=4,cex=2.25,xaxt="n",xlab="")
# grid()
# abline (0,1,col=2, lwd=2)
# abline(h=0,v=0,lty=3,lwd=2)
# points (S_nopooling, S_shrunk, pch=4,lwd=3, col=bmmb::cols,cex=2.25)
plot (L_nopooling, L_shrunk-L_nopooling, xlim = c(-10,10), ylim = c(-5,5),
pch=3, lwd=3, col=rev(bmmb::cols),cex=2.25,cex.lab=0.9,cex.axis = 0.9)
grid()
abline(h=0,v=0,lty=3,lwd=2)
points (L_nopooling, L_shrunk-L_nopooling, pch=3,lwd=3, col=bmmb::cols,cex=2.25)
mtext (side = 1, text = "L_nopooling", line=2.5,cex=0.9)
plot (S_nopooling, S_shrunk-S_nopooling, xlim = c(-12,12), ylim = c(-5,5),
pch=3, col=4,cex=2.25,cex.lab=0.9,cex.axis = 0.9)
grid()
abline(h=0,v=0,lty=3,lwd=2)
points (S_nopooling, S_shrunk-S_nopooling, pch=3,lwd=3, col=rev(bmmb::cols),cex=2.25)
mtext (side = 1, text = "S_nopooling", line=2.5,cex=0.9)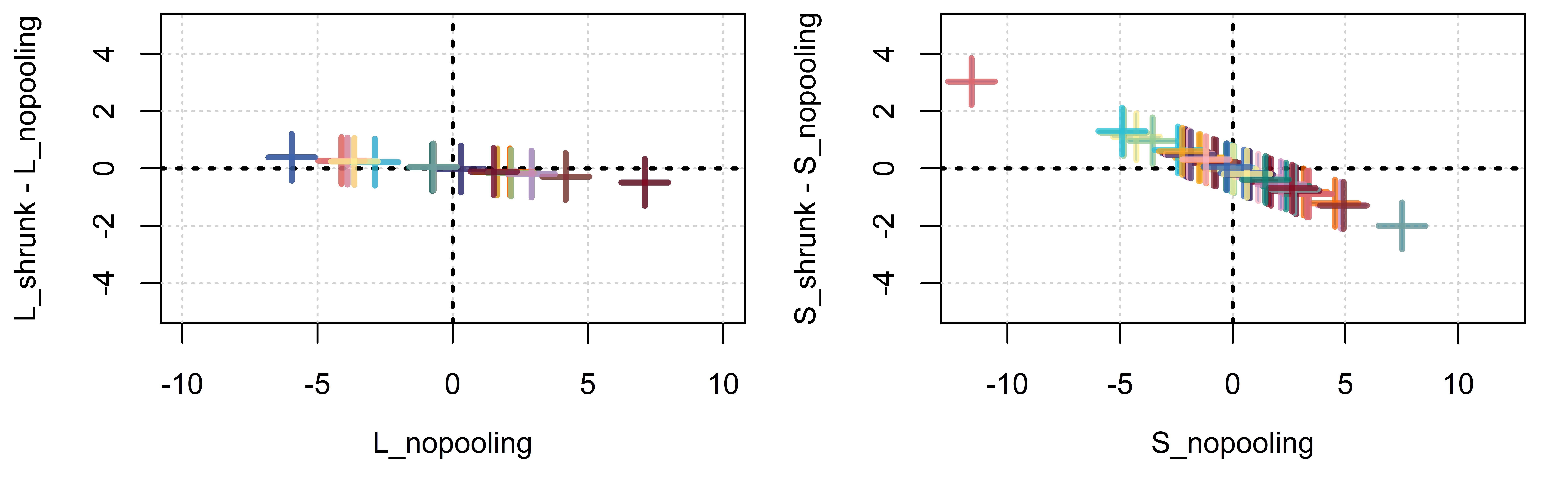
You may have noted that the listener effects exhibit less shrinkage than the speaker effects. There are two reasons for this. First, we have 45 observations for each listener but only 15 for each speaker. As a result, the likelihood functions for our listener effects will be narrower than those of our speaker effects. This makes the likelihoods for our listener parameters more resistant to the effects of the prior, and therefore more resistant to shrinkage (see sections Section 3.3.2 and Section 2.7.3) for more information). Second, a large proportion of the speaker effects are close to zero, with only a relatively small number exhibiting large deviations from this. In contrast, the listener effects are less concentrated near the average. As a result of this \(\sigma_L > \sigma_S\), which causes speaker effects to be shrunken more than listener effects, in general.
4.10 Answering our research questions
Let’s return to the research questions we posed in Section 4.5.1:
(Q1) How tall does the average adult male sound?
(Q2) Can we set limits on credible average apparent heights based on the data we collected?
We can consider the answers provided to this question by the complete pooling model we fit at the end of last chapter:
bmmb::short_summary (model_priors)
## Formula: height ~ 1
## Population-Level Effects:
## Estimate Est.Error l-95% CI u-95% CI
## Intercept 173.8 0.31 173.2 174.4
##
## Family Specific Parameters:
## Estimate Est.Error l-95% CI u-95% CI
## sigma 7.77 0.21 7.37 8.19And compare this to the final model we considered above, which contained information about repeated measurements across speakers and listeners:
bmmb::short_summary (model_multilevel_L_S)
## Formula: height ~ 1 + (1 | L) + (1 | S)
##
## Group-Level Effects:
## ~L (Number of levels: 15)
## Estimate Est.Error l-95% CI u-95% CI
## sd(Intercept) 3.81 0.86 2.51 5.87
##
## ~S (Number of levels: 45)
## Estimate Est.Error l-95% CI u-95% CI
## sd(Intercept) 2.83 0.42 2.1 3.72
##
## Population-Level Effects:
## Estimate Est.Error l-95% CI u-95% CI
## Intercept 173.8 1.12 171.6 176.1
##
## Family Specific Parameters:
## Estimate Est.Error l-95% CI u-95% CI
## sigma 6.47 0.19 6.11 6.85Our complete pooling model from the previous chapter, model_priors, and our adaptive pooling model from this chapter, model_multilevel_L_S, agree on the average apparent height as reflected by their respective intercept terms. However, they disagree on a credible interval for the parameter, with the complete pooling model having a 95% credible interval that is nearly four times narrower (1.2 cm vs 4.4 cm). This is because the complete pooling model did not specify information about repeated measures, which caused our model to think that it had more independent observations than it did. This led the initial model to return an overly-precise estimate.
Another difference is that the final model has a smaller sigma parameter (7.8 vs 6.5), which indicates that the data-level error is smaller in the final model than in the initial model. Keep in mind that ‘error’ is just what your model can’t explain at the data level. Our final model explains more (at the listener and speaker levels) and so there is less error (at the data level). The reduced error is a direct consequence of the fact that the final model splits variation in the data into several components (i.e. \(\sigma^2_{total}=\sigma^2_{L}+\sigma^2_{S}+\sigma^2\)). Since \(\sigma_{total}\) is a fixed value (given the data), obviously, the larger your between-speaker and between-listener variation is, the smaller your random error (\(\sigma\)) must be.
Usually, parameters should be reported with at least the mean/median and standard deviations of the posterior distribution, in addition to some useful credible interval (i.e. 50%, 95%) around that parameter. Based on the result of our final model, a thorough answer to our research questions might go something like:
Based on our model the average apparent height of adult males is likely to be 173.8 cm (s.d. = 1.1, 95% CI = [171.6, 176.1]). The estimated magnitude of the random error was 6.5 cm (s.d. = 0.2, 95% CI = [6.1, 6.9]). Systematic between-listener variation averages about 3.8 cm (s.d. = 0.9, 95% CI = [2.5, 5.8]), while systematic between-speaker variation averages about 2.8 cm (s.d. = 0.4, 95% CI = [2.1, 3.8]).
4.11 Traditionalist’s corner
In traditionalist’s corner, we’re going to compare the output of brms to some more ‘traditional’ approaches. We’re not going to talk about the traditional models in any detail, the focus of this section is simply to highlight the similarities between different approaches and to point out where to find equivalent information in the different models. If you are already familiar with these approaches, these sections may be helpful. If not, some of the information provided here may not make much sense, although it may still be helpful. If you want to know more about the statistical methods being discussed here, please see the preface for a list of suggested background reading in statistics.
4.11.1 Bayesian multilevel models vs. lmer
Here we compare the output of brms to the output of the lmer (“linear mixed-effects regression”) function, a very popular function for fitting multilevel models in the lme4 R package. We’re not going to talk about lmer in very much detail. The focus of this section is simply to highlight the similarities between different approaches, and to point out where to find equivalent information in both kinds of models. Below we use lmer to fit a model that is analogous to our model_multilevel_L_S model. The lmer function uses adaptive pooling to estimate its ‘random effects’ and no pooling to estimate its ‘fixed effects’. Notice that the formulas are identical in both cases, except we do not specify priors for this model.
lmer_model = lme4::lmer (height ~ 1 + (1|L) + (1|S), data = men)We can consider the short print statement for our Bayesian model, which we have annotated with numbers indicating where certain information can be found.
bmmb::short_summary(model_multilevel_L_S)
## (1) Formula: height ~ 1 + (1 | L) + (1 | S)
##
## (2) Group-Level Effects:
## ~L (Number of levels: 15)
## Estimate Est.Error l-95% CI u-95% CI
## (3) sd(Intercept) 3.81 0.86 2.51 5.87
##
## ~S (Number of levels: 45)
## Estimate Est.Error l-95% CI u-95% CI
## (4) sd(Intercept) 2.83 0.42 2.1 3.72
##
## Population-Level Effects:
## Estimate Est.Error l-95% CI u-95% CI
## (5) Intercept 173.8 1.12 171.6 176.05
##
## Family Specific Parameters:
## Estimate Est.Error l-95% CI u-95% CI
## (6) sigma 6.47 0.19 6.11 6.85And compare this to the print statement of our lmer model, annotated with the same numbers highlighting equivalent information. We see that our models provide very similar information, although it is organized (and named) in somewhat different ways. We also see that the two approaches provide us with very similar parameter estimates for our models.
summary (lmer_model)
## Linear mixed model fit by REML ['lmerMod']
## (1) Formula: height ~ 1 + (1 | L) + (1 | S)
## Data: men
##
## REML criterion at convergence: 4527.4
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -4.6205 -0.4868 0.0722 0.5700 2.7179
##
## (2) Random effects:
## Groups Name Variance Std.Dev.
## (3) S (Intercept) 7.593 2.756
## (4) L (Intercept) 11.990 3.463
## (6) Residual 41.630 6.452
## Number of obs: 675, groups: S, 45; L, 15
##
## Fixed effects:
## Estimate Std. Error t value
## (5) (Intercept) 173.788 1.015 171.3We can get the random effects out of our brm model and compare them to the random effects we get from lmer. As seen below, the random effects estimates we get from brm also include credible intervals. As a result, we have some idea regarding the uncertainty in these estimates.
brms_ranefs = brms::ranef (model_multilevel_L_S)$L[,,"Intercept"]
head (brms_ranefs)
## Estimate Est.Error Q2.5 Q97.5
## 1 -3.6312 1.346 -6.366 -1.07315
## 2 -2.6594 1.371 -5.350 0.01293
## 3 1.5297 1.368 -1.158 4.23982
## 4 -0.7165 1.358 -3.380 1.95702
## 5 -3.8413 1.361 -6.472 -1.18510
## 6 3.8990 1.353 1.266 6.58467In contrast, lmer gives you what are called point estimates. These are single estimates of parameter values with no intervals indicating uncertainty. Because of this, it is difficult to compare estimates for different speakers/participants in the data.
lmer_ranefs = lme4::ranef (lmer_model)[["L"]]
head (lmer_ranefs)
## (Intercept)
## 1 -3.6134
## 2 -2.6726
## 3 1.5298
## 4 -0.7024
## 5 -3.8238
## 6 3.8796Importantly, the values we get from both approaches are nearly identical, as seen in Figure 4.10.
show plot code
################################################################################
brms_ranefs_S = brms::ranef (model_multilevel_L_S)$S[,,"Intercept"]
lmer_ranefs_S = lme4::ranef (lmer_model)[["S"]]
par(mfrow = c(1,2), mar = c(4,4.5,1,1))
brmplot (brms_ranefs, xlab="Listener", ylab="Listener Effect (cm)", col=deepgreen, line = FALSE)
abline (h = 0)
points (lmer_ranefs[,1],pch=4,cex=1.5,lwd=2,col=coral, xaxt='n',xlab='',
ylab="")
brmplot (brms_ranefs_S, xlab="Speaker", ylab="Speaker Effect (cm)", col=deepgreen, line = FALSE)
abline (h = 0)
points (lmer_ranefs_S[,1],pch=4,cex=1.5,lwd=2,col=coral, xaxt='n',xlab='',
ylab="")
#plot (brms_ranefs[,1], lmer_ranefs[,1], lwd=2, cex=1.5, col=deeppurple,
# xlab = 'brms Random Effects (cm)', ylab = 'lmer Random Effects (cm)', pch=16)
#abline (0,1,col=2)
#abline (h = 0, v = 0, lty=3)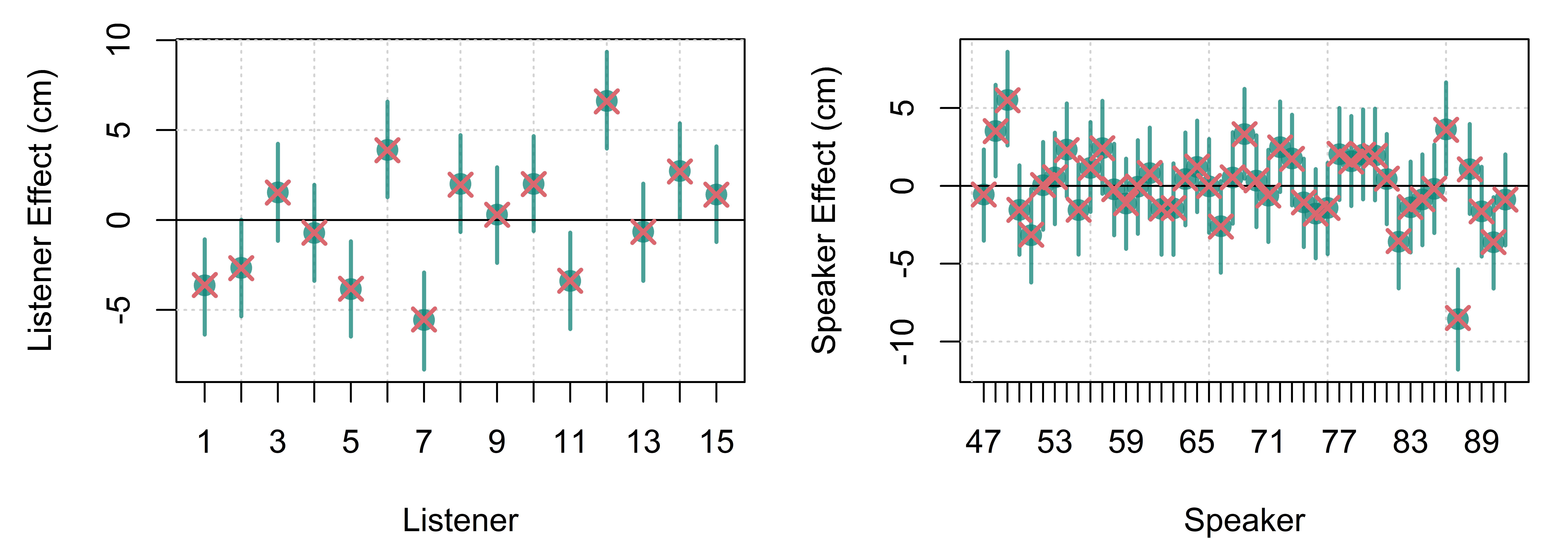
The largest difference between the two sets of coefficients is 0.05 cm for the listener effects and 0.06 cm for the speaker effects, with the average absolute difference for both being around 0.03 cm. So, as a practical matter analyzing this data using a Bayesian multilevel model provides some advantages (e.g. intervals around random effects), while still providing effectively the same ‘answers’ as a ‘traditional’ approach to the data.
4.12 Exercises
The analyses in the main body of the text all involve only the unmodified ‘actual’ resonance level (in exp_data). Responses for the stimuli with the simulated ‘big’ resonance are reserved for exercises throughout. You can get the ‘big’ resonance in the exp_ex data frame, or all data in the exp_data_all data frame.
Fit and interpret one or more of the suggested models:
Easy: Analyze the (pre-fit) model that’s exactly like
model_multilevel_L_S, except using the data inexp_ex(bmmb::get_model("model_multilevel_L_S_ex.RDS")).Medium: Fit a model just like
model_multilevel_L_S, but for the data from some other group, for either the original or big resonance levels.Hard: Fit two models like
model_multilevel_L_Sfor two groups, or for one group across resonance levels, and compare results across models.
In any case, describe the model, present and explain the results, and include some figures.
4.13 References
Bates D, Mächler M, Bolker B, Walker S (2015). “Fitting Linear Mixed-Effects Models Using lme4.” Journal of Statistical Software, 67(1), 1–48. doi:10.18637/jss.v067.i01.
Gelman, A., & Hill, J. (2006). Data analysis using regression and multilevel/hierarchical models. Cambridge university press.
Gelman, A., Carlin, J. B., Stern, H. S., Dunson, D. B., Vehtari, A., & Rubin, D. B. (2013). Bayesian Data Analysis.
4.14 Plot Code
################################################################################
library (bmmb)
data (exp_data)
men = exp_data[exp_data$C_v=='m',]
# create vector of height judgments
mens_height = men$height
#par (mar = c(.1,.1,.5,.1), mfrow = c(1,1), oma = c(4.2,.20,0,.2))
#layout (m=matrix(c(1,2,3,4),2,2), heights = c(.25,.75))
par (mar = c(4,.5,.5,.5), mfrow = c(1,2))
#plot (0,type='n',xlab='',ylab='',xaxt='n',yaxt='n',bty='n')
boxplot (men$height,col = 'grey', ylim = c(140,195),
width = 2, ylab="", horizontal = TRUE)
mtext (side=2,outer=TRUE, text="Apparent height (cm)", cex=1, line=2.9,adj=.55)
#boxplot (tapply(men$height,men$L,mean), xlab = "",xaxt='n',
# col = cols, ylim = c(140,195), width = 2,cex.axis=1.3,
# horizontal = TRUE)
#abline (h=)
#text (1,190,expression(sigma["[within]"]), cex=2)
boxplot (height ~ L, data = men, xlab = "", ylab="",xaxt='s',
col = cols[-1], ylim = c(140,195), cex.lab=1.1,cex.axis=1.1,
horizontal = TRUE, yaxt='n')
grid()
abline (h = mean(mens_height), lwd=2, lty=3)
boxplot (height ~ L, data = men, col = cols[-c(1,8)],add=TRUE,yaxt='n',xaxt='n')
mtext (side=1,outer=TRUE, "Apparent height (cm)",line=-0.9)
################################################################################
knitr::include_graphics("images/Figure 4.2.jpg")
################################################################################
par (mfrow = c(1,1), mar = c(4,4,0.5,0.5))
boxplot (height ~ L, data = men, main = "",xlab="Listener",
ylab="Apparent height (cm)",
col=bmmb::cols, ylim = c(150,195), outline=FALSE)
abline (h = mean(men$height), lwd=3,lty=3)
boxplot (height ~ L, data = men, main = "",col=bmmb::cols,add=TRUE, outline=FALSE)
################################################################################
par (mfrow = c(1,1), mar = c(4,4,1,1))
height_centered = (men$height-mean(men$height))
boxplot (height_centered ~ L, data=men, col = 0, ylim = c(-23,21), xlab="Listener",
ylab="Centered apparent height (cm)", outline = FALSE)
abline (h=0)
bmmb::brmplot(listener_intercepts, col = bmmb::cols, ylim = c(-30,20),
add = TRUE, outline = FALSE)
################################################################################
par (mfrow = c(1,2), mar = c(0.5,0.5,1,0.5), oma = c(0,3.5,0,0))
boxplot (height_rep ~ L, ylim = c(140,200), xaxt='n',xlab='',
col=bmmb::cols)
abline (h = 173.8, lwd=3,lty=3)
boxplot (height ~ L, data = men, ylim = c(140,200), xaxt='n',xlab='',
col=bmmb::cols,yaxt="n")
abline (h = 173.8, lwd=3,lty=3)
mtext (text = "Apparent height (cm)", side=2, outer=TRUE, line = 2.5)
################################################################################
par (mfrow = c(3,1), mar = c(.1,3,.1,1), oma = c(1,2,1,0))
boxplot (height_rep_1 ~ L, ylim = c(140,198),xaxt='n',
col=bmmb::cols,ylab="",yaxt = 'n')
text (1, 145, label = "Intercept + error", cex = 1.5, pos=4)
abline (h=174.8,lty=2)
axis (side=2,at=seq(150,190,10))
boxplot (height_rep_2 ~ L, ylim = c(140,198),xaxt='n',
col=bmmb::cols,ylab="",yaxt = 'n')
abline (h=174.8,lty=2)
text (1, 145, label = "Intercept + L", cex = 1.5, pos=4)
axis (side=2,at=seq(150,190,10))
boxplot (height_rep ~ L, ylim = c(140,198),xaxt='n',
col=bmmb::cols,ylab="",yaxt = 'n')
abline (h=174.8,lty=2)
text (1, 145, label = "Intercept + L + error", cex = 1.5, pos=4)
mtext (side=2,text="Apparent height (cm)", outer = TRUE, line=0)
axis (side=2,at=seq(150,190,10))
################################################################################
library (bmmb)
data (exp_data)
men = exp_data[exp_data$C_v=='m',]
# create vector of height judgments
mens_height = men$height
#par (mar = c(.1,.1,.5,.1), mfrow = c(1,1), oma = c(4.2,.20,0,.2))
#layout (m=matrix(c(1,2,3,4),2,2), heights = c(.25,.75))
par (mar = c(4.5,.5,.5,.5), mfrow = c(1,3))
#plot (0,type='n',xlab='',ylab='',xaxt='n',yaxt='n',bty='n')
boxplot (men$height,col = 'grey', ylim = c(140,195),
width = 2, ylab="", horizontal = TRUE)
mtext (side=2,outer=TRUE, text="Apparent height (cm)", cex=1, line=2.9,adj=.55)
#boxplot (tapply(men$height,men$L,mean), xlab = "",xaxt='n',
# col = cols, ylim = c(140,195), width = 2,cex.axis=1.3,
# horizontal = TRUE)
#abline (h=)
#text (1,190,expression(sigma["[within]"]), cex=2)
boxplot (height ~ L, data = men, xlab = "", ylab="",xaxt='s',
col = cols[-1], ylim = c(140,195), cex.lab=1.1,cex.axis=1.1,
horizontal = TRUE, yaxt='n')
grid()
abline (h = mean(mens_height), lwd=2, lty=3)
boxplot (height ~ L, data = men, col = cols[-c(1,8)],add=TRUE,yaxt='n',xaxt='n')
boxplot (height ~ S, data = men, xlab = "", ylab="",xaxt='s',
col = cols[-1], ylim = c(140,195), cex.lab=1.1,cex.axis=1.1,
horizontal = TRUE, yaxt='n')
grid()
abline (h = mean(mens_height), lwd=2, lty=3)
boxplot (height ~ S, data = men, col = cols[-c(1,8)],add=TRUE,yaxt='n',xaxt='n')
mtext (side=1,outer=TRUE, "Apparent height (cm)",line=-1.3)
################################################################################
par (mfrow = c(1,2), mar = c(2,.51,.5,.51))
layout (mat = matrix(c(1,3,2,3), 2,2))
plot (L_nopooling,rep(0,15), ylim = c(0,4.3), cex=1.5,pch=1,col=rev(bmmb::cols), xlim = c(-13,13),
yaxt = 'n',xlab = 'Listener Effects',lwd=3)
points (L_shrunk,rep(1,15), cex=1.5,pch=16,col=rev(bmmb::cols),lwd=2)
arrows (L_nopooling,0.1,L_shrunk,0.9, length=0.1)
abline (v=0, lty=3,col='grey')
x = seq(-12,12,0.01)
den = dnorm (x, 0, 3.79)
den = den / max (den)
den2 = dnorm (x, 0, sd(S_nopooling))
den2 = den2 / max (den2)
text (0,2.5,label = expression(paste(sigma["L"]," = 3.79")), cex = 1.25)
text (0,3,label = expression(paste("P(L)")), cex = 1.25)
lines (x, (den*2)+2, lwd = 3, col = 4)
#lines (x, (den2*2)+2, lwd = 2, col = 4,lty=3)
text (-13.5,0,"No pooling", pos = 4)
text (-13.5,1,"Shrunken", pos = 4)
text (5,4,"Listener Effects", pos = 4)
plot (S_nopooling,rep(0,45), ylim = c(0,4.3), cex=1.5,pch=1,col=rev(bmmb::cols), xlim = c(-13,13),
yaxt = 'n', ylab = '',xlab = 'Speaker Effects',lwd=3)
points (S_shrunk,rep(1,45), cex=1.5,pch=16,col=rev(bmmb::cols),lwd=2)
arrows (S_nopooling,0.1,S_shrunk,0.9, length=0.1)
abline (v=0, lty=3,col='grey')
x = seq(-12,12,0.01)
den = dnorm (x, 0, 2.84)
den = den / max (den)
den2 = dnorm (x, 0, sd(L_nopooling))
den2 = den2 / max (den2)
text (0,2.5,label = expression(paste(sigma["S"]," = 2.84")), cex = 1.25)
text (0,3,label = expression(paste("P(S)")), cex = 1.25)
lines (x, (den*2)+2, lwd = 3, col = 4)
#lines (x, (den2*2)+2, lwd = 2, col = 4,lty=3)
text (5,4,"Speaker Effects", pos = 4)
text (-12, 0.5, "S=87")
plot (S_nopooling[41],0.05, ylim = c(0,1.5), cex=2.5,pch=1,
col=bmmb::cols[5], xlim = c(-17,9), yaxt = 'n', ylab = '',
xlab = 'Speaker Effects',lwd=3, yaxs = 'i')
points (S_shrunk[41],0.05, cex=2.5,pch=16,col=bmmb::cols[5],lwd=2)
arrows (S_nopooling[41],0.05,S_shrunk[41]-.5,0.05, length=0.1)
x = seq(-20,15,0.01)
den = dnorm (x, 0, 2.84)
den = den / max (den)
tmp = exp_data$height[exp_data$S==87] - 173.8
den2 = dnorm (x, mean (tmp), 6.47/sqrt(14))
den2 = den2 / max (den2)
den3 = den2 * den
den3 = den3 / max (den3)
den4 = dnorm (x, -8.560 , 1.648)
den4 = den4 / max (den4)
use2 = (x > -17) & (x < -5)
lines (x[use2], (den2)[use2], lwd = 3, col = bmmb::cols[5], lty = 2)
#lines (x[use2], (den4)[use2], lwd = 3, col = bmmb::cols[6], lty = 2)
use3 = (x > -13) & (x < -2)
lines (x[use3], (den3[use3]), lwd = 3, col = bmmb::cols[5])
use = (x > -10) & (x < 10)
lines (x[use], (den[use]), lwd = 3, col = 4)
points (S_nopooling[41],0.05, cex=2.5,pch=1,col=bmmb::cols[5])
points (S_shrunk[41],0.05, cex=2.5,pch=16,col=bmmb::cols[5],lwd=2)
text (S_nopooling[41],1.25,label = expression(paste("P(y|",S["[87]"],")")), cex = 1.25)
text (S_shrunk[41],1.25,label = expression(paste("P(",S["[87]"],"|y)")), cex = 1.25)
text (0,1.25,label = expression(paste("P(",S,")")), cex = 1.25)
text (S_nopooling[41],.5,label = "Likelihood", cex = 1.25)
text (S_shrunk[41],.5,label = "Posterior", cex = 1.25)
text (0,.5,label = "Prior", cex = 1.25)
abline (v=0,lty=3,col="grey")
################################################################################
par (mfrow = c(1,2), mar = c(.25,4,.25,1), oma=c(4,.1,.1,.1))
#
# plot (L_nopooling, L_shrunk, xlim = c(-10,10), ylim = c(-10,10),
# pch=4, lwd=3, col=bmmb::cols,cex=2.25,xaxt="n",xlab="")
# grid()
# abline (0,1,col=2,lwd=2)
# abline(h=0,v=0,lty=3,lwd=2)
# points (L_nopooling, L_shrunk, pch=4,lwd=3, col=bmmb::cols,cex=2.25)
#
# plot (S_nopooling, S_shrunk, xlim = c(-12,12), ylim = c(-12,12),
# pch=4, col=4,cex=2.25,xaxt="n",xlab="")
# grid()
# abline (0,1,col=2, lwd=2)
# abline(h=0,v=0,lty=3,lwd=2)
# points (S_nopooling, S_shrunk, pch=4,lwd=3, col=bmmb::cols,cex=2.25)
plot (L_nopooling, L_shrunk-L_nopooling, xlim = c(-10,10), ylim = c(-5,5),
pch=3, lwd=3, col=rev(bmmb::cols),cex=2.25,cex.lab=0.9,cex.axis = 0.9)
grid()
abline(h=0,v=0,lty=3,lwd=2)
points (L_nopooling, L_shrunk-L_nopooling, pch=3,lwd=3, col=bmmb::cols,cex=2.25)
mtext (side = 1, text = "L_nopooling", line=2.5,cex=0.9)
plot (S_nopooling, S_shrunk-S_nopooling, xlim = c(-12,12), ylim = c(-5,5),
pch=3, col=4,cex=2.25,cex.lab=0.9,cex.axis = 0.9)
grid()
abline(h=0,v=0,lty=3,lwd=2)
points (S_nopooling, S_shrunk-S_nopooling, pch=3,lwd=3, col=rev(bmmb::cols),cex=2.25)
mtext (side = 1, text = "S_nopooling", line=2.5,cex=0.9)
################################################################################
brms_ranefs_S = brms::ranef (model_multilevel_L_S)$S[,,"Intercept"]
lmer_ranefs_S = lme4::ranef (lmer_model)[["S"]]
par(mfrow = c(1,2), mar = c(4,4.5,1,1))
brmplot (brms_ranefs, xlab="Listener", ylab="Listener Effect (cm)", col=deepgreen, line = FALSE)
abline (h = 0)
points (lmer_ranefs[,1],pch=4,cex=1.5,lwd=2,col=coral, xaxt='n',xlab='',
ylab="")
brmplot (brms_ranefs_S, xlab="Speaker", ylab="Speaker Effect (cm)", col=deepgreen, line = FALSE)
abline (h = 0)
points (lmer_ranefs_S[,1],pch=4,cex=1.5,lwd=2,col=coral, xaxt='n',xlab='',
ylab="")
#plot (brms_ranefs[,1], lmer_ranefs[,1], lwd=2, cex=1.5, col=deeppurple,
# xlab = 'brms Random Effects (cm)', ylab = 'lmer Random Effects (cm)', pch=16)
#abline (0,1,col=2)
#abline (h = 0, v = 0, lty=3)